Summary
Maintenance of body temperature in cold-exposed animals requires induction of thermogenesis and management of fuel. Here, we demonstrated that reducing ambient temperature attenuated diet-induced obesity (DIO), which was associated with increased iBAT thermogenesis and a plasma bile acid profile similar to that of germ-free mice. We observed a marked shift in the microbiome composition at the phylum and family levels within 1 day of acute cold exposure and after 4 weeks at 12°C. Gut microbiota was characterized by increased levels of Adlercreutzia, Mogibacteriaceae, Ruminococcaceae, and Desulfovibrio and reduced levels of Bacilli, Erysipelotrichaceae, and the genus rc4-4. These genera have been associated with leanness and obesity, respectively. Germ-free mice fed a high-fat diet at room temperature gained less adiposity and improved glucose tolerance when transplanted with caecal microbiota of mice housed at 12°C compared to mice transplanted with microbiota from 29°C. Thus, a microbiota-liver-BAT axis may mediate protection against obesity at reduced temperature.
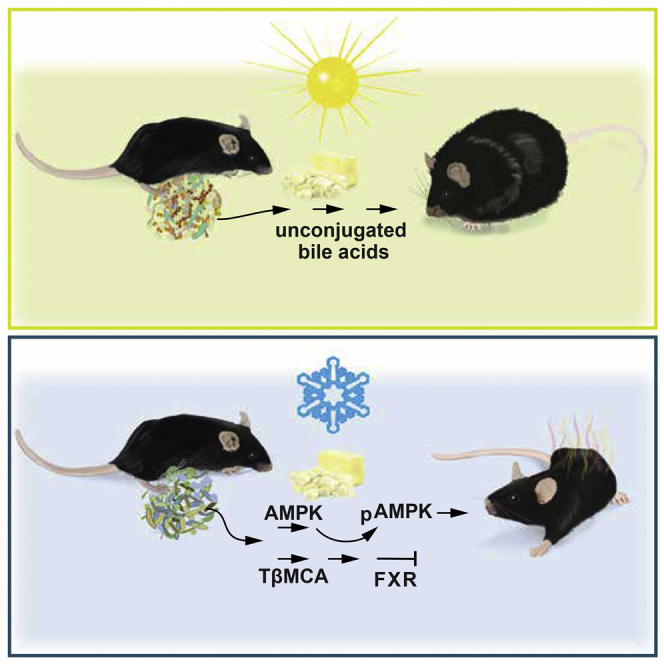
Graphical Abstract
Highlights
-
•
Gut microbiota is altered during suppression of obesity in a cold environment
-
•
Cold-adapted microbiota improves host bile acid and energy metabolism
-
•
Acute effects of cold-adapted microbiota are linked to thermogenesis in iBAT
-
•
Cold-adapted microbiota may signal through FXR and AMPK
Zietak et al. show that the gut microbiota contributes to the beneficial effects of cold on adiposity via a mechanism involving the production of conjugated bile acids and AMPK activation. Transfer of gut microbiota from cold-exposed mice reduces diet-induced obesity in germ-free recipient mice.
Introduction
Regulation of body weight and adiposity relies on a homeostatic system balancing energy intake with energy expenditure. Well-documented evidence gathered during the past 35 years has established that induction of thermogenesis in brown adipocytes of mice and rats can reduce obesity (Rothwell and Stock, 1979). The maintenance of body temperature by brown adipose tissue (BAT) thermoregulation utilizes energy stores in proportion to ambient temperature (Kozak, 2010). Importantly, BAT-based non-shivering thermogenesis may be important in humans as well (Yoneshiro and Saito, 2015). Therefore, adiposity may be regulated by the capacity of the individual to manipulate brown adipocyte phenotypes at reduced temperature, thereby constituting a strategy to reduce metabolic efficiency (Jaroslawska et al., 2015).
During the past decade, accumulating data have demonstrated that the gut microbiota impacts body weight and energy homeostasis (Tremaroli and Bäckhed, 2012). Microbial diversity as well as the relative proportions between the members of main phyla Firmicutes and Bacteroidetes have been associated with regulation of obesity in both mice and humans (Ley et al., 2005, Ley et al., 2006). The gut microbiota is important for energy harvest on a polysaccharide-rich diet (Bäckhed et al., 2004) and also contributes to diet-induced obesity (DIO) by modulating different pathways involving triglyceride storage and fatty acid oxidation (Bäckhed et al., 2007). Interestingly, microbiota from lean twins can protect against obesity induced by microbiota from obese twins, when transferred to mice (Ridaura et al., 2013).
Bile acids (BAs) produced by hepatocytes, stored in the gall bladder and released into the gut after a meal, are metabolized by the gut microbiota to generate an array of BA species such as secondary BAs, e.g., lithocholic and deoxycholic acid (Sayin et al., 2013). These can activate nuclear receptor farnesoid X receptor (FXR) and G-coupled receptor TGR5, located on iBAT, to regulate thermogenesis (Prawitt et al., 2011b, Watanabe et al., 2006). Recent studies have linked changes in the gut microbiota in a cold environment to the thermogenic potential of brite cells through enhanced type 2 cytokine signaling by the innate immune system (Lee et al., 2015, Chevalier et al., 2015). Here we show, in contrast to the role of brite cells in determining resistance to DIO in C57BL/6J mice at reduced temperature (Suárez-Zamorano et al., 2015), that acute changes in energy balance in mice fed a high-fat diet at 12°C is associated with changes in the gut microbiota, increased BA metabolism, and induction of iBAT thermogenesis.
Results
Reduced Ambient Temperature Protects against DIO by Inducing Thermogenesis
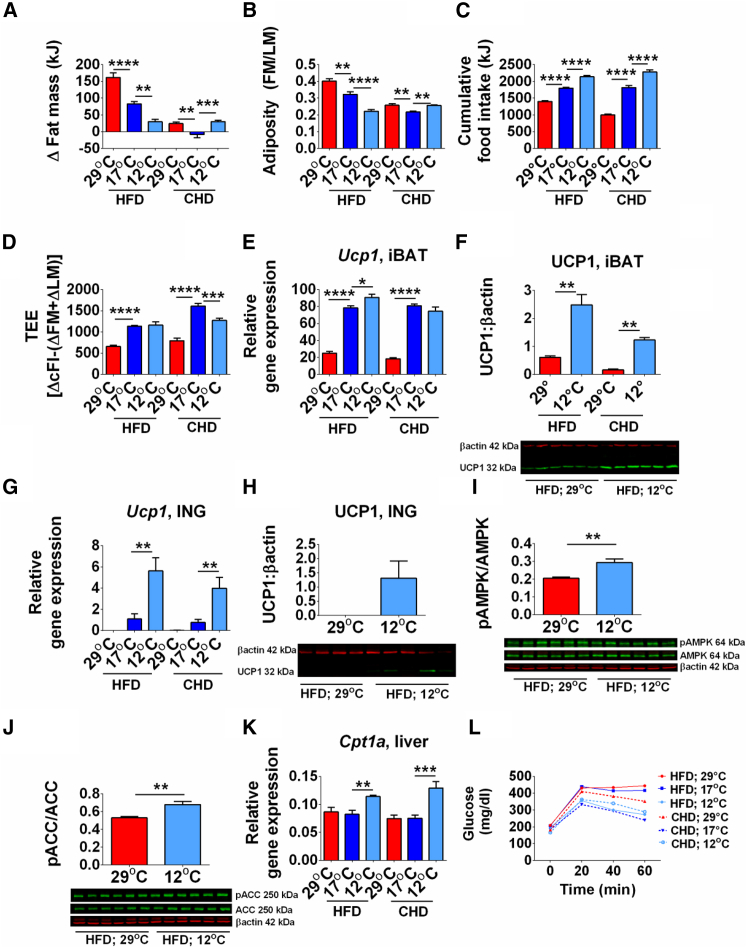
We fed mice with high-fat diet (HFD) and chow diet (CHD) at thermoneutrality and at reduced ambient temperatures to assess the effects of reduced temperature on cold-induced energy expenditure and development of DIO. At 12°C and 17°C, the reduced fat mass and adiposity (Figures 1A, 1B, S1A, and S1B) together with increased food intake in mice fed either HFD or CHD resulted in increased energy expenditure (Figures 1C and 1D), demonstrating that reduced temperature blocks DIO. Furthermore, energy balance is maintained by increasing food intake.
Figure 1.
Energy Balance Phenotype in Response to Reduced Ambient Temperature and Thermoneutrality in Mice Fed HFD or CHD for 4 Weeks
(A–D) Change in fat mass (A); adiposity index (B); cumulative food intake (C); total energy expenditure (D).
(E–H) Ucp1 mRNA (E) and protein (F) by immunoblot in iBAT. Ucp1 mRNA (G) and protein (H) by immunoblot in ING.
(I–K) Hepatic induction of pAMPK (I), pACC (J), and Cpt1a mRNA level (K).
(L) Glucose tolerance test.
Data are presented as mean ± SEM. n = 8–10 (GTT and qRT-PCR) or 5–6 (immunoblot). FM, fat mass; LM, lean mass; cFI, cumulative food intake; TEE, total energy expenditure. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S1.
Reduction of ambient temperature from 29°C to 17°C and 12°C induced iBAT and inguinal fat (ING) uncoupling protein 1 (Ucp1) mRNA and protein expression in mice fed either CHD or HFD (Figures 1E–1H and S1C). However, Ucp1 expression in ING was only 6% of that observed in iBAT. In addition, Pgc1a (peroxisome proliferator-activated receptor coactivator 1-alpha) and Adrb1 (beta3-adrenergic receptor) expression, which are associated with improved metabolism, was significantly induced in iBAT and ING by cold (Figures S1D and S1E). Reduced cellular energy levels lead to increased phosphorylation of AMPK and was associated with increased phosphorylation of ACC and increased expression of Cpt1a, indicating increased hepatic fatty acid oxidation (Figures 1I–1K). At 12°C mice had decreased expression of biomarkers of hepatic lipogenesis (Figure S1F), a phenotype associated with reduced plasma cholesteryl esters (CEs) and triacylglycerols (TAGs) in mice fed HFD at 12°C compared to 29°C (CEs, 12°C 6,102.5 ± 446 versus 29°C 8,421.1 ± 490.7, p < 0.05; TAGs, 12°C 155.1 ± 50.4 versus 29°C 409.4 ± 43.4, p < 0.05).
Modulation of Glucose Tolerance and Metabolism by Ambient Temperature
Consistent with reduced adiposity, glucose tolerance was improved in mice fed HFD at 12°C compared to 17°C or 29°C and was similar in both HFD- and CHD-fed mice at 12°C (Figure 1L). Reduced ambient temperature also improved insulin tolerance of mice on either HFD or CHD (Figures S1G and S1H). The improved glucose tolerance was associated with increased Glut4 (glucose transporter type 4) mRNA and protein levels in iBAT and skeletal muscle (Figures S1I–S1K). Induction of Irs1 (insulin receptor substrate 1) mRNA at 17°C reflected the improved insulin tolerance at reduced ambient temperature (Figure S1L). Furthermore, expression of Pepck (phosphoenolpyruvate carboxykinase) and G6pc (glucose-6-phosphatase catalytic subunit) mRNA levels was increased in liver of mice housed at 17°C and 12°C (Figures S1M and S1N), indicating increased hepatic gluconeogenesis, which provides energy for thermogenesis in the cold (Himms-Hagen, 1995).
Diet and Ambient Temperature Alter Gut Microbiota Composition
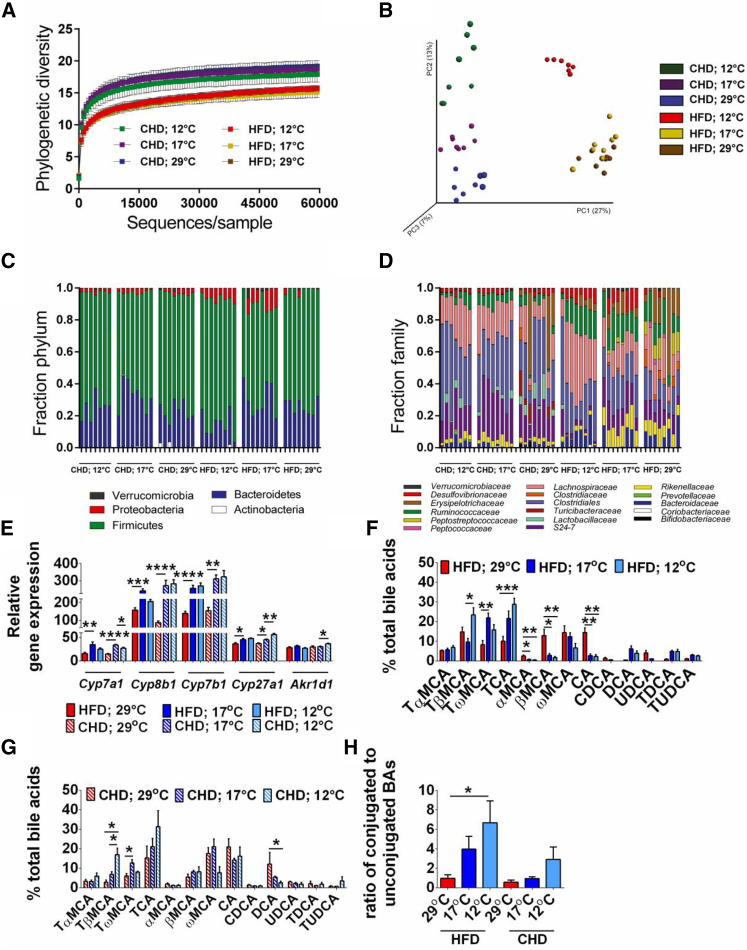
To directly investigate the impact of diet and ambient temperature on mouse gut microbiota, the variable region 4 (V4) of bacterial 16S rRNA genes was amplified by PCR and sequenced using the Illumina MiSeq platform. We observed 4,090 distinct OTUs (from 9,442,775 reads; ranging from 75,344 to 159,323 reads per sample). To investigate how diet and ambient temperature affected the microbiota phylogenetic richness in each caecum sample, we analyzed the α-diversity, as assessed by rarefication and phylogenetic diversity (Figure 2A). Similar to previous reports, we found that diet is a key factor in shaping phylogenetic diversity (Hildebrandt et al., 2009, Turnbaugh et al., 2009) and observed that the caecal microbiota of mice on CHD had higher phylogenetic diversity compared with mice on HFD. We next performed principal coordinate analysis (PCoA) of unweighted UniFrac distances between the caecum samples from the different groups to determine the effects of diet and ambient temperature on the microbiota. A clear separation between the communities, driven by diet, was observed at the first principal coordinate (x axis), which explained 27% of the variance (Figure 2B). The second principal coordinate (y axis) accounted for 13% of the variance and separated the communities by ambient temperature within the diets (Figure 2B). Moreover, the communities of mice maintained at 12°C separated from those at 17°C and 29°C independently of the diets with the strongest effect in the HFD groups (Figure 2B).
Figure 2.
The Gut Microbiota Composition and Microbial Metabolism Shaped by Ambient Temperature and Diet
(A–H) Rarefaction curves calculated for phylogenetic distance between the microbiota of mice fed a HFD and a CHD at 12°C, 17°C, and 29°C. Rarefaction depth used is 60,000 sequences per sample (A). Principal coordinate analysis (PCoA) of unweighted UniFrac revealed clustering of the gut microbiota after diet and temperatures. The percentage of the variation explained by the plotted principal coordinates is indicated in the axis labels. Each dot represents a caecal community (B). Relative abundance at phylum (C) and family (D) level in caecal community of mice fed a HFD and a CHD at 12°C, 17°C, and 29°C. Hepatic expression of genes involved in BA synthesis (E). Plasma BA profile in mice fed a HFD (F), a CHD (G), and a ration of t-conjugated to unconjugated BAs (H). The data are given as mean ± SEM. n = 8. p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S2.
Analysis at the phylum level indicated that the caecal microbiota was dominated by five major phyla: Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia (Figure 2C). Mice fed HFD at 12°C and 17°C were associated with a bloom of Proteobacteria and a reduction in Bacteroidetes and an increase in Firmicutes (Figure 2C). Shifts in Bacteroidaceae, Rikenellaceae, and S24-7 (all Bacteroidetes), particularly in S24-7 on CHD and Bacteroidaceae on HFD, were observed (Figure 2D). The increase in Firmicutes under all conditions was due to increases in Clostridiaceae, Lachnospiraceae, and Ruminococcaceae (Figure 2D). Interestingly, a bloom in Erysipelotrichaceae was observed in both diets, but only at 29°C (Figure 2D). The bloom in Proteobacteria at reduced temperature was largely accounted for by Desulfovibrionaceae (Figure 2D), which has been previously associated with metabolic health (Caesar et al., 2015).
To identify taxonomic differences in the microbiota composition of mice fed HFD or CHD and challenged by differences in the ambient temperature and to identify specific bacterial taxa or species-level phylotypes that contribute to the adiposity phenotype, we applied linear discriminant analysis (LDA) effect size (LEfSe) with LDA score > 2 (Segata et al., 2011). This analysis revealed 46 discriminative features in the CHD-fed mice (Figure S2A) and 34 in the microbiota of HFD-fed mice (Figure S2B). Similar OTUs belonging to class Bacilli in particular orders Turicibacterales and Lactobacillales, the genus Allobaculum in the family Erysipelotrichaceae, as well as the genus rc4-4 in the family Peptococcaceae (Figures S2A and S2B) were increased in mice maintained at 29°C independent of diet. The microbiota of mice fed HFD maintained at 12°C was enriched in a Corriobacteria such as Adlercreutzia, number of Clostridia from the Mogibacteriaceae and Ruminococcaceae families, and Desulfovibrionales such as Desulfovibrio (Figure S2B). Similar changes were observed in mice fed CHD maintained at 17°C (Figure S2A), all of which were lean and glucose tolerant.
Reduced Ambient Temperature and Diet Regulate Bile Acid Metabolism
Reduced ambient temperature caused a robust induction of hepatic enzymes engaged in the conversion of cholesterol to primary BAs, including CYP7A1 (cholesterol 7alpha-hydroxylase), CYP8B1 (sterol 12alpha-hydroxylase), and CYP27A1 (sterol 27-hydroxylase), as well as in taurine production and taurine conjugation to BAs (Figures 2E, S2C, and S2D). Accordingly, the BA profile was drastically altered and dominated by unconjugated BA (CA, and α-, βMCA) at 29°C in mice fed a HFD, whereas primary taurine-conjugated BAs, i.e., TCA, TαMCA, and TβMCA, were elevated in plasma of mice maintained at 12°C on both diets (Figures 2F–2H).
FGF21 was 7- and 2.5-fold upregulated in iBAT at 12°C on a HFD and CHD, respectively (Figure S2E). Furthermore, type 2 iodothyronine deiodinase expression, which is associated with increased thermogenesis, was induced at reduced temperature (Figure S2E), as previously observed (de Jesus et al., 2001). The TGR5 receptor, which binds to microbially produced secondary BAs (Kawamata et al., 2003), was induced at 12°C while thyroid hormone receptor beta was not (Figures S2E–S2G).
Gut Microbiota Composition Is Determined by Reduced Ambient Temperature
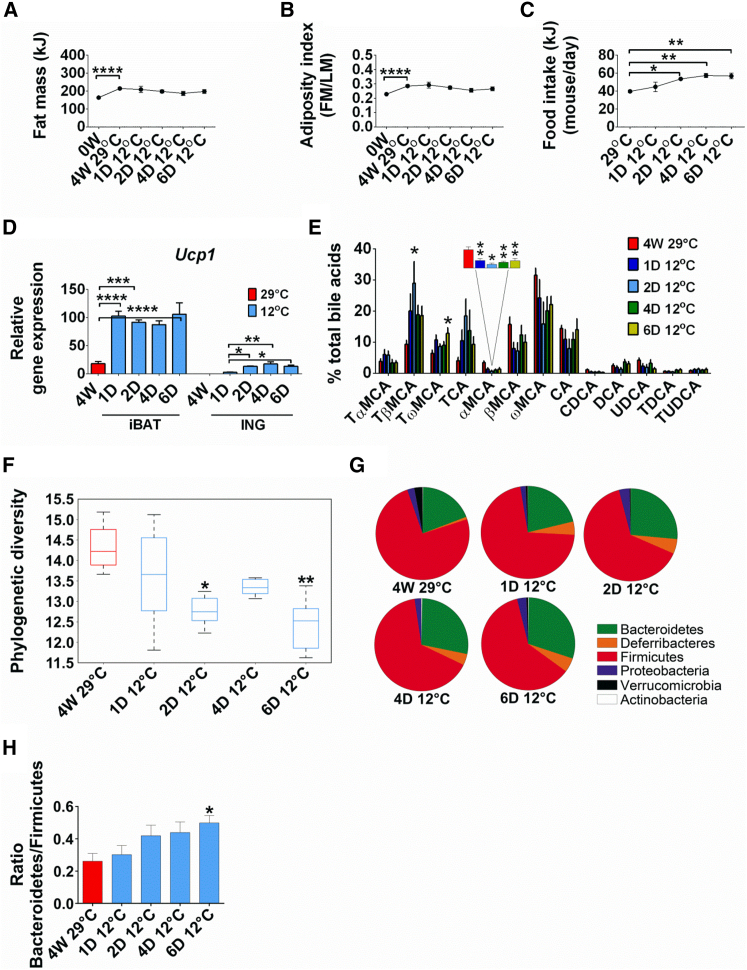
To investigate whether the gut microbiota and BAs were altered in response to reduced temperature or reduction in obesity, we performed a kinetic experiment and analyzed the microbiota in mice that were fed HFD for 4 weeks at 29°C and then transferred to 12°C for 6 days. Increased fat mass and adiposity after 4 weeks of a HFD at 29°C was not affected by reduced ambient temperature (Figures 3A and 3B), and the increased demand for fuel was satisfied by a 20% increase in food intake within 2 days of cold exposure (Figure 3C). Cold exposure induced Ucp1 expression 5-fold in iBAT after 1 day at 12°C (Figure 3D), similar to the increase observed after 4 weeks at 12°C (compare with Figure 1E). The induction of Ucp1 in ING was very low and therefore not associated with the initial response required for survival in the cold (Figure 3D).
Figure 3.
Time-Dependent Effects of Acute Cold Exposure on Energy Balance Phenotypes and Gut Microbiota Composition of Mice with Diet-Induced Obesity
(A–H) Fat mass (A), adiposity (B), food intake (C), and Ucp1 expression in iBAT and ING (D) of mice fed a HFD and housed at 29°C for 4 weeks, then transferred to 12°C for 6 days. Change in BA composition measured in plasma (E), phylogenetic diversity comparison of caecal microbial communities (F), phylum level abundance (G), and ratio of Bacteroidetes to Firmicutes (H) of the microbiota of mice with DIO exposed to 6 days of cold exposure. Data are presented as mean ± SEM or ±SD (graph F). n = 6. ∗p < 0.05; ∗∗p < 0.01. FM, fat mass; LM, lean mass. See also Figure S3.
The gut microbiota and BA composition were shifted to the cold-adapted state within 1 day, demonstrating that these changes were independent of reduced adiposity (Figures 3E–3H). The BAs profile, already modified 1 day after cold exposure, was dominated by conjugated BAs (TCA, TαMCA, TβMCA, and TωMCA), whereas the levels of unconjugated BAs (αMCA, βMCA, and ωMCA) were reduced compared to 29°C (Figure 3E). Analysis on caecal microbiota composition showed a significant decrease in the phylogenetic diversity already 2 days after cold exposure compared to 29°C (Figure 3F). At the phylum level, Deferribacteres increased and Verrucomicrobia decreased within 1 day of cold (Figure 3G). Bacteroidetes increased and Firmicutes decreased, but not until after 6 days at 12°C (Figure 3G), to increase the Bacteroidetes/Firmicutes ratio (Figure 3H). At the family level, the most abundant phylotypes affected by cold exposure were Bacteroidaceae, Rikenellaceae, Porphyromonadaceae, Deferribacteraceae, Lactobacillaceae, Clostridiaceae, and Peptostreptococcaceae (Figure S3A). The fraction of others, such as S24-7, Clostridiales, Lachnospiraceae, Ruminococcaceae, Erysipelotrichaceae, and Desulfovibrionaceae (Figure S3A) did not change when mice were transferred to cold for 6 days, suggesting that these microbial phylotypes are associated with HFD feeding. Similar to the initial cold-exposure experiment for 4 weeks, we observed that Adlercreutzia and Bacteroides were increased after 6 days of cold exposure (Figure S3B). The Firmicutes, Lactobacillus, Clostridiaceae, genus SMB53, Dehalobacterium, Peptostreptococcaceae, and Mogibacteriaceae decreased at 12°C (Figure S3B). Long-term cold exposure induced a bloom in Proteobacteria, particularly in Desulfovibrionaceae (Figure S2B), but it did not change during the short-term exposure at 12°C (Figure S3A). These data suggest that in parallel with Ucp1 induction, changes in microbiota and BAs occur when ambient temperature is reduced, but independent of changes in adiposity.
The Gut Microbiota Suppresses the Development of DIO at Reduced Ambient Temperature
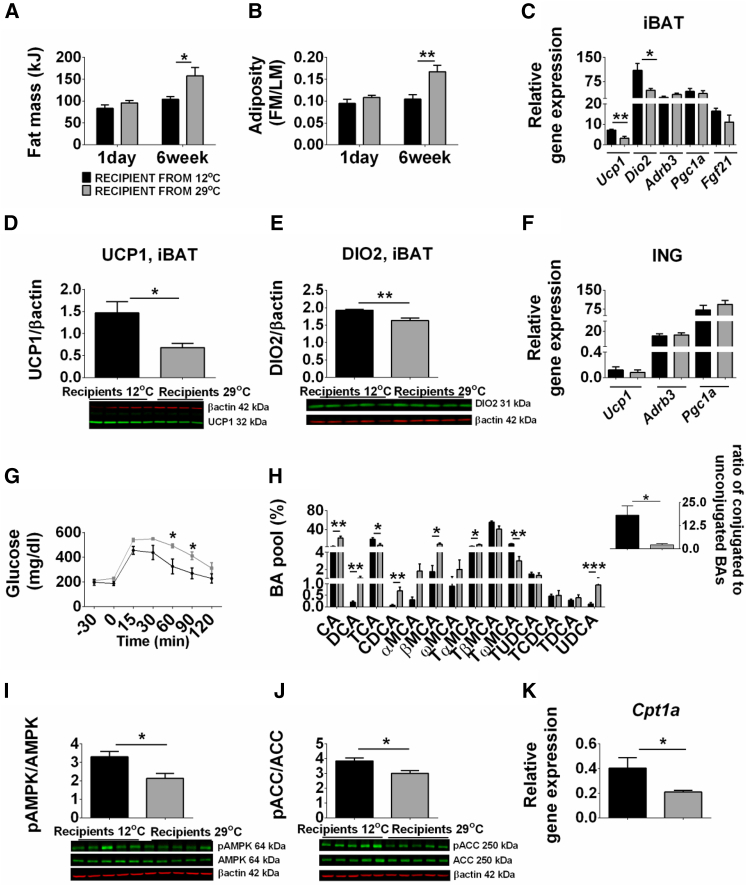
To assess the capacity of the gut microbiota from mice maintained at reduced ambient temperature to modulate the development of DIO, we performed a microbiota transplantation experiment. Mice colonized with microbiota from donors kept at 12°C had reduced fat mass and adiposity as well as significantly higher expression of Ucp1 and Dio2 mRNA and protein in iBAT compared with mice colonized with microbiota from 29°C (Figures 4A–4E). Expression of Adrb3, Pgc1a, and Fgf21was not altered in iBAT (Figure 4C); no changes in gene expression were detected in ING of recipient mice (Figure 4F). The glucose response was improved in the recipient mice colonized with microbiota from donor kept at 12°C, but no differences occurred in GLUT4 protein levels in muscle and iBAT (Figures 4G, S4A, and S4B). Recipient mice, colonized with microbiota from donors kept at 12°C, had increased hepatic expression of Cyp8b1 and Cyp7b1 (25-hydroxycholesterol 7alpha-hydroxylase) and Csd (cysteine sulfinic acid decarboxylase), leading to an altered BA profile (Figures S4C and 4H). These mice had an increased proportion of conjugated, and a reduced proportion of primary unconjugated, BAs (CA, CDCA, and βMCA) and secondary BAs (DCA and UDCA) in the plasma (Figure 4H). Gut microbiota transferred from donors housed at 12°C influenced hepatic lipid metabolism, inducing AMPK phosphorylation augmenting lipid β-oxidation in liver compared with microbiota from 29°C (Figures 4I–4K and S4D). Nonetheless, lipid analysis showed a modest increase in total TAG in plasma in mice colonized with microbiota from donor kept at 12°C (Figure S4E). These results suggest that functional differences between microbiota of mice kept at 12°C and 29°C can be transferred to recipient mice kept at 23°C.
Figure 4.
Transfer of Cold-Adapted Microbiota Improves Metabolic Phenotype
(A–J) Fat mass (A), adiposity index (B), and changes in mRNA levels of thermogenic genes (C) and protein levels of UCP1 (D) and DIO2 (E) in iBAT and mRNA levels of thermogenic genes in ING (F) in GF mice colonized with microbiota harvested from mice kept at 12°C or 29°C. Glucose tolerance test (G). BA profile of recipient mice (H). pAMPK (I) and pACC (J) protein levels in liver and hepatic Cpt1a (K) expression of recipient mice. The data are given as mean ± SEM. n = 4–5 per group. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. FM, fat mass; LM, lean mass. See also Figure S4.
Discussion
A primary and essential task of all endothermic animals is to maintain a normal body temperature by inducing thermogenesis in response to cold. Here, we demonstrate that reducing ambient temperature from 29°C to 12°C or even 17°C also protects the host from DIO, an effect that is associated with increased Ucp1 expression in iBAT. However, recent studies have shown that the pathway for activating thermogenesis may be more complex than the traditional model based on iBAT thermogenesis. Thermogenesis occurs not only in iBAT, but also by the induction of brite cells in WAT at 29°C by administration of interleukin 4 (Qiu et al., 2014). Similarly, the thermogenic program in iBAT may be activated by BAs at 29°C (Zietak and Kozak, 2016). Recently, it was shown that the depletion of the microbiota by antibiotic treatment or GF conditions stimulates the induction of brite cells by enhanced type 2 cytokine signaling from eosinophil infiltration (Suárez-Zamorano et al., 2015). Here, we show that both the acute and chronic exposure to reduced ambient temperature leads to induction of Ucp1 in iBAT and rapid changes in the composition of the gut microbiota and BA metabolism. The results suggest that the induction of cold-mediated thermogenesis involves the activation of iBAT thermogenesis by a mechanism involving modulation of BA metabolism and AMPK phosphorylation by the gut microbiota.
A key question is whether the improved obesity/diabetic phenotype of mice with DIO reared in the cold is dependent on the effects of the gut microbiota. We observed that cold exposure is linked to altered caecal microbiota and that mice on both diets shared microbial phylotypes associated with changes in ambient temperature. Bacteria belonging to Bacilli and Erysipelotrichaceae, usually associated with obesity (Turnbaugh et al., 2009), were enriched in mice at 29°C, whereas Adlercreutzia and Desulfovibrio, associated with leanness (Caesar et al., 2015, Goodrich et al., 2014), were increased at 12°C. Changes in the composition of gut microbiota occurring within 1 day of exposure at 12°C and in the absence of changes in adiposity suggest that the changes in microbiota are driven by changes in temperature rather than merely reflecting obesity. The major microbiota changes occurring in parallel with induction of Ucp1 were reduced Firmicutes apparent after 1 day and an increase in Bacteroidetes after 6 days, changes previously linked with protection against obesity (Ley et al., 2005, Turnbaugh et al., 2008). Consistent with the study of Chevalier et al. (2015), we identified a decreased abundance of Verrucomicrobia and an increased abundance of Deferribacteres when mice were exposed to cold, suggesting that altered abundance of members in these phyla contribute to the cold-induced phenotype. Changes in other bacteria after longer cold exposure may be in response to altered adiposity of the host.
The mechanism by which the gut microbiota influences adiposity may include modulation of BA metabolism (Sayin et al., 2013). BAs are microbial-derived metabolic regulators, which can suppress DIO through increased energy expenditure (Watanabe et al., 2012). Here we show that lower ambient temperature increased production of BAs and expression of genes related to BA synthesis. The increased levels of conjugated BAs may be attributed to reduced levels of Lactobacillus, which has high deconjugation capacity upon cold exposure. These observations were confirmed in the kinetics study where conjugated BAs were increased and Lactobacillus was decreased after 1 day at 12°C. The increased prevalence of taurine species in the cold might antagonize FXR receptor (Sayin et al., 2013) and protect against DIO as FXR-deficient mice are resistant to DIO (Prawitt et al., 2011a). In agreement with inhibition of FXR signaling in the cold-exposed animals, we observed increased expression of enzymes involved in BA synthesis and increased BA pool size. The cold-exposed mice had a BA profile that resembled that of GF mice, which also are resistant to DIO and have increased expression of phosphorylated AMPK in the liver (Bäckhed et al., 2007). AMPK activation inhibits FXR activity (Lien et al., 2014) and also promotes energy expenditure. Accordingly, we observed increased expression of phosphorylated ACC as well as downstream Cpt1a expression, which is associated with increased fatty acid oxidation and energy expenditure. Importantly, we observed this phenotype both following cold-exposure as well as after microbiota transfer, suggesting that the phenotype is linked to the altered microbiota.
In the current study, cold exposure at 12°C causes a strong and rapid induction of the brown adipose phenotype in iBAT, but not in ING, an increased whole-animal energy expenditure, and a complete suppression of DIO.
We demonstrated that transplantation of caecal material from mice reared at 12°C to GF recipients improved their metabolic phenotype. Consistent with our findings, Chevalier et al. (2015) demonstrated that transplantation of caecal material from cold-exposed mice reduced obesity and improved insulin sensitivity. In contrast to our findings, they emphasized browning in ING WAT. However, the brown adipocyte phenotype in ING WAT in our study is minor (1%–5%) compared to the robust iBAT thermogenic phenotype.
Our investigations suggest that changes in the gut microbiota in response to the cold exposure mediate BA metabolism, possibly through changes in AMPK and FXR signaling, to complement sympathetic signaling in the regulation of thermogenesis in iBAT and resistance to DIO.
Experimental Procedures
Animals and Study Design
Protocol I. Breeding pairs of C57BL6/J (B6) mice were obtained from the Jackson Laboratory. Adult B6 mice at 12–20 weeks of age were divided into 6 groups (n = 8–10 mice/group). Three groups were ad libitum fed HFD (58% energy in kcal from fat, AIN-76A 9G03 Research Diets); another three groups received CHD (11% energy in kcal from fat, 5053, rodent diet 20, LabDiet). Mice were single-housed with a 12 hr light/12 hr dark cycle at environmental temperatures of 12°C, 17°C, and 29°C for 4 weeks.
Protocol II. B6 mice at 8 weeks of age were individually housed at 29°C and ad libitum fed a HFD for 4 weeks (n = 6 mice). Thereafter, mice were transferred to 12°C for 1, 2, 4, and 6 days and received ad libitum a HFD (n = 6 mice/time point).
For both protocols, mice were anaesthetized with an overdose of cocktail composed of ketamine/xylazine/chlorpromazine administered subcutaneously to collect blood samples by cardiac puncture. Blood samples were centrifuged for 10 min at 2,400 rpm and stored at −20°C. Mice were sacrificed by cervical dislocation, and tissues and caecal content were quickly removed and stored at −80°C for further analysis. Animal experiments performed at different temperatures were approved by the Local Committee for the Ethical Treatment of Experimental Animals of Warmia-Mazury University (NR 38/2011), Olsztyn, Poland.
Phenotyping
Fat mass and lean mass were measured by nuclear magnetic resonance (NMR, Bruker). Energy content of fat mass and lean mass were expressed in kJ. Adiposity index was determined from body composition. Body weight was measured weekly. Food intake was calculated on a weekly basis and expressed as the amount of energy in kJ. At the end of the experiment, glucose and insulin tolerance tests were performed.
Bacterial Genomic DNA Isolation
Bacterial gDNA was extracted using the GeneMATRIX Stool DNA Purification Kit (EURx) from the caecum of mice fed HFD and maintained at 17°C or 29°C. Genomic DNA from caecum of mice fed HFD at 12°C and CHD at 12°C, 17°C, and 29°C was isolated using the repeated bead beating (RBB) method (Salonen et al., 2010). Details about 16S rRNA amplification, sequencing, microbiota data analysis, and BA analyses are provided in Supplemental Experimental Procedures.
Statistical Analysis
The data are expressed as mean ± SEM and analyzed using GraphPad Prism 6.0. Statistical differences for single variables were analyzed by Mann-Whitney test or one-way ANOVA and Tukey’s multiple comparison post hoc test for three groups (29°C, 17°C, and 12°C) The level of significance was set at p < 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Author Contributions
M.Z. was involved in design of the experiment and performed physiological and molecular phenotypes and participated in writing the manuscript; P.K.-D. was involved in design and performed experiments concerning microbial profiling and transplant experiments and participated in writing the manuscript; L.H.M. and M.S. participated in experimentation and discussion; L.P.K. designed the initial hypothesis and participated in the design and the writing of the manuscript; F.B. was involved in design and interpretation of experiments concerning microbial profiling and transplant experiments and participated in the design and the writing of the manuscript.
Acknowledgments
We thank Anna Hallén for excellent assistance with the transfer experiments and for producing the graphical abstract. 16S rRNA sequencing was performed at the Genomic Core Facility of University of Gothenburg and in the F.B. lab with the assistance of Manuela Krämer. Preprocessing of sequencing data was performed by Rozita Akrami and Valentina Tremaroli. This work was supported by a grant to L.P.K. from the Foundation for Polish Science, programme WELCOME, no. WELCOME/2010-4/3 entitled “Nutrition and ambient temperature during early development can reduce susceptibility to obesity” financed by EU Structural Funds in Poland within the Innovative Economy Programme and REFRESH project FP7-REGPOT-2010-1-264103). This study was also supported by the Swedish Research Council, Torsten Söderberg’s foundations, Göran Gustafsson’s foundation, Novo Nordisk Foundation, Swedish Foundation for Strategic Research, Knut and Alice Wallenberg foundation, and the regional agreement on medical training and clinical research (ALF) between Region Västra Götaland and Sahlgrenska University Hospital. F.B. is a recipient of ERC Consolidator Grant (European Research Council, Consolidator grant 615362 - METABASE). F.B. is a founder and shareholder of Metabogen AB.
Published: June 14, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.cmet.2016.05.001.
Contributor Information
Leslie P. Kozak, Email: l.kozak@pan.olsztyn.pl.
Fredrik Bäckhed, Email: fredrik.backhed@wlab.gu.se.
Accession Numbers
The accession number for the bacteria sequencing data reported in this paper is Sequence Read Archive: PRJNA321010.
Supplemental Information
References
- Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F., Manchester J.K., Semenkovich C.F., Gordon J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar R., Tremaroli V., Kovatcheva-Datchary P., Cani P.D., Bäckhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier C., Stojanović O., Colin D.J., Suarez-Zamorano N., Tarallo V., Veyrat-Durebex C., Rigo D., Fabbiano S., Stevanović A., Hagemann S. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163:1360–1374. doi: 10.1016/j.cell.2015.11.004. [DOI] [PubMed] [Google Scholar]
- de Jesus L.A., Carvalho S.D., Ribeiro M.O., Schneider M., Kim S.W., Harney J.W., Larsen P.R., Bianco A.C. The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue. J. Clin. Invest. 2001;108:1379–1385. doi: 10.1172/JCI13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J.K., Waters J.L., Poole A.C., Sutter J.L., Koren O., Blekhman R., Beaumont M., Van Treuren W., Knight R., Bell J.T. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt M.A., Hoffmann C., Sherrill-Mix S.A., Keilbaugh S.A., Hamady M., Chen Y.Y., Knight R., Ahima R.S., Bushman F., Wu G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137 doi: 10.1053/j.gastro.2009.08.042. 1716–24.e1, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himms-Hagen J. Role of brown adipose tissue thermogenesis in control of thermoregulatory feeding in rats: a new hypothesis that links thermostatic and glucostatic hypotheses for control of food intake. Proc. Soc. Exp. Biol. Med. 1995;208:159–169. doi: 10.3181/00379727-208-43847a. [DOI] [PubMed] [Google Scholar]
- Jaroslawska J., Chabowska-Kita A., Kaczmarek M.M., Kozak L.P. Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status. PLoS Genet. 2015;11:e1005287. doi: 10.1371/journal.pgen.1005287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata Y., Fujii R., Hosoya M., Harada M., Yoshida H., Miwa M., Fukusumi S., Habata Y., Itoh T., Shintani Y. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- Kozak L.P. Brown fat and the myth of diet-induced thermogenesis. Cell Metab. 2010;11:263–267. doi: 10.1016/j.cmet.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.W., Odegaard J.I., Mukundan L., Qiu Y., Molofsky A.B., Nussbaum J.C., Yun K., Locksley R.M., Chawla A. Activated type 2 innate lymphoid cells regulate beige fat biogenesis. Cell. 2015;160:74–87. doi: 10.1016/j.cell.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Bäckhed F., Turnbaugh P., Lozupone C.A., Knight R.D., Gordon J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Lien F., Berthier A., Bouchaert E., Gheeraert C., Alexandre J., Porez G., Prawitt J., Dehondt H., Ploton M., Colin S. Metformin interferes with bile acid homeostasis through AMPK-FXR crosstalk. J. Clin. Invest. 2014;124:1037–1051. doi: 10.1172/JCI68815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt J., Abdelkarim M., Stroeve J.H., Popescu I., Duez H., Velagapudi V.R., Dumont J., Bouchaert E., van Dijk T.H., Lucas A. Farnesoid X receptor deficiency improves glucose homeostasis in mouse models of obesity. Diabetes. 2011;60:1861–1871. doi: 10.2337/db11-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prawitt J., Caron S., Staels B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr. Diab. Rep. 2011;11:160–166. doi: 10.1007/s11892-011-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Nguyen K.D., Odegaard J.I., Cui X., Tian X., Locksley R.M., Palmiter R.D., Chawla A. Eosinophils and type 2 cytokine signaling in macrophages orchestrate development of functional beige fat. Cell. 2014;157:1292–1308. doi: 10.1016/j.cell.2014.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura V.K., Faith J.J., Rey F.E., Cheng J., Duncan A.E., Kau A.L., Griffin N.W., Lombard V., Henrissat B., Bain J.R. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell N.J., Stock M.J. A role for brown adipose tissue in diet-induced thermogenesis. Nature. 1979;281:31–35. doi: 10.1038/281031a0. [DOI] [PubMed] [Google Scholar]
- Salonen A., Nikkilä J., Jalanka-Tuovinen J., Immonen O., Rajilić-Stojanović M., Kekkonen R.A., Palva A., de Vos W.M. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods. 2010;81:127–134. doi: 10.1016/j.mimet.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Sayin S.I., Wahlström A., Felin J., Jäntti S., Marschall H.U., Bamberg K., Angelin B., Hyötyläinen T., Orešič M., Bäckhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Zamorano N., Fabbiano S., Chevalier C., Stojanović O., Colin D.J., Stevanović A., Veyrat-Durebex C., Tarallo V., Rigo D., Germain S. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015;21:1497–1501. doi: 10.1038/nm.3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Bäckhed F., Fulton L., Gordon J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ridaura V.K., Faith J.J., Rey F.E., Knight R., Gordon J.I. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Houten S.M., Mataki C., Christoffolete M.A., Kim B.W., Sato H., Messaddeq N., Harney J.W., Ezaki O., Kodama T. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439:484–489. doi: 10.1038/nature04330. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Morimoto K., Houten S.M., Kaneko-Iwasaki N., Sugizaki T., Horai Y., Mataki C., Sato H., Murahashi K., Arita E. Bile acid binding resin improves metabolic control through the induction of energy expenditure. PLoS ONE. 2012;7:e38286. doi: 10.1371/journal.pone.0038286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneshiro T., Saito M. Activation and recruitment of brown adipose tissue as anti-obesity regimens in humans. Ann. Med. 2015;47:133–141. doi: 10.3109/07853890.2014.911595. [DOI] [PubMed] [Google Scholar]
- Zietak M., Kozak L.P. Bile acids induce uncoupling protein 1-dependent thermogenesis and stimulate energy expenditure at thermoneutrality in mice. Am. J. Physiol. Endocrinol. Metab. 2016;310:E346–E354. doi: 10.1152/ajpendo.00485.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.