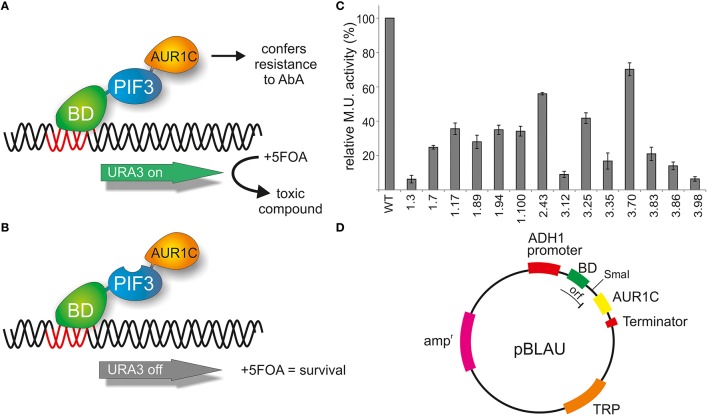
Figure 1.
Reverse yeast-1-hybrid scheme for TAD identification. (A) PIF3 is fused N-terminally to the DNA binding domain (BD) of the yeast GAL4 transcription factor and C-terminally to AUR1C, which confers resistance to AbA. AUR1C serves as a second selectable marker, providing verification of the expression of the fusion construct, and thus reducing false positives, such as premature stop codons, frameshift mutations, or low-expression clones. The PIF3 transcription factor self-activates transcription of the reporter gene URA3 in yeast. URA3 converts 5FOA (Fluroorotic acid) into a toxic compound, leading to negative selection. (B) In the screen, randomly mutagenized PIF3 sequences were inserted between BD and AUR1C. If the mutagenized PIF3 fails to activate transcription, URA3 will not be expressed, leading to survival in the presence of 5FOA. (C) Relative transcriptional activity (rel. Miller units) of 14 randomly selected colonies identified with the rY1H method. (D) Schematic representation of the rY1H vector pBLAU. The coding sequences of BD and AUR1C markers are out of frame, to reduce background of intact pBLAU plasmid under screening conditions. Insertion of the protein of interest into the SmaI opened vector pBLAU is done via homologous recombination. The reading frame of the fusion protein is restored by adding an additional base in the reverse primer used for homologous recombination.