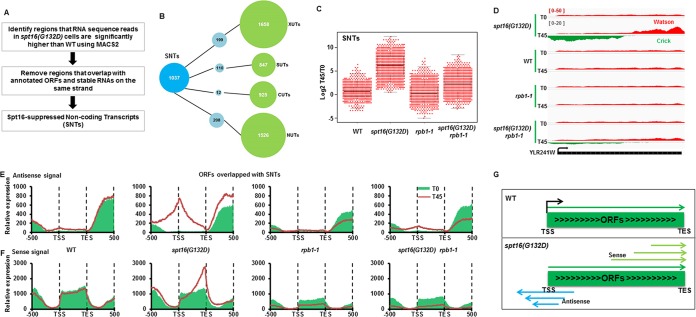
FIG 4.
Inactivation of Spt16 results in bidirectional cryptic transcription. (A) Schematic diagram for the identification of Spt16-suppressed noncoding transcripts (SNTs). (B) Overlap of SNTs with other previously annotated noncoding RNAs, cryptic unstable transcripts (CUTs) and stable unannotated transcripts (SUTs) from Xu et al. (38), Xrn1-sensitive unstable transcripts (XUTs) from van Dijk et al. (39), and Nrd1 unterminated transcripts (NUTs) from Schulz et al. (69). If an SNT shares more than 50% length with each known ncRNA, it would be counted as one that overlaps with the known ncRNAs. (C) SNTs are produced by RNA polymerase II. To compare the expression of each SNT in the WT, spt16(G132D), rpb1-1, and spt16(G132D) rpb1-1 mutant cells, the log2 ratio of the RPKM of the individual SNTs at 37°C for 45 min (T45) over 0 min (T0) [log2(T45/T0)] was calculated and plotted using a dot-box plot. Each dot represents one SNT. (D) An SNT overlapping with YLR241W shows increased SNT transcripts in the spt16(G132D) mutant cells at T45 compared to T0 in both the sense and antisense directions. RNA-seq reads for the Watson strand and Crick strand are shown. (E and F) Both antisense and sense noncoding transcripts increase at ORFs that overlapped SNTs. The yeast ORFs were separated into two groups based on whether they overlapped an SNT. Antisense (E) and sense (F) of RNA-Seq reads of ORFs that overlapped SNTs were calculated and plotted from 500 bp upstream TSS to 500 bp downstream TES. The results for ORFs without overlapped SNTs are presented in Fig. S7 in the supplemental material. Red (T0, 25°C) and green lines (T45, 37°C) represent treatments at two different temperatures. (G) A model for the generation of noncoding transcripts through bidirectional cryptic transcription in the spt16(G132D) mutant cells. It is likely that SNT initiates transcription at multiple start sites and thus generates the SNT transcripts shown in panels E and F.