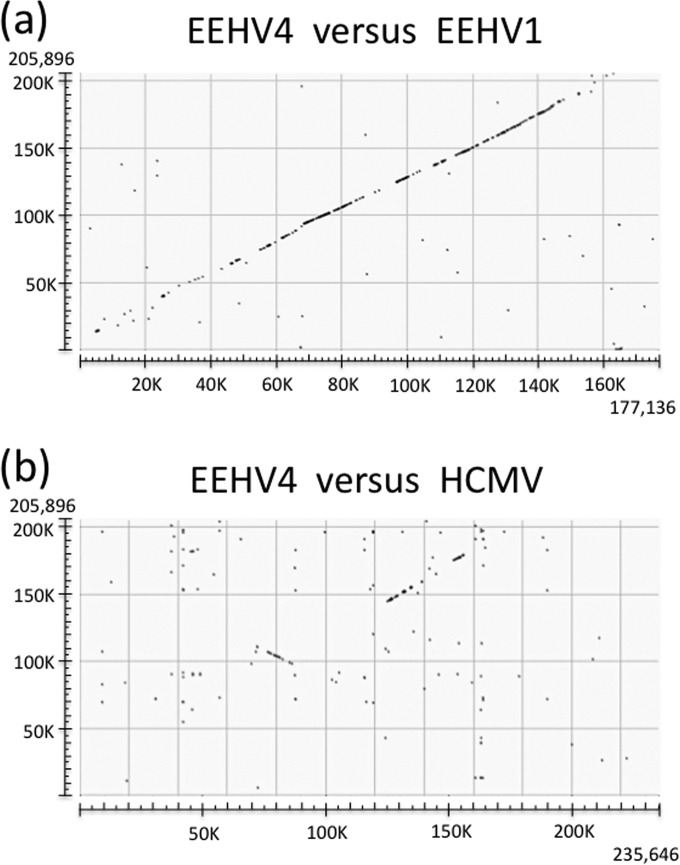
FIG 2 .
Global alignment patterns for the intact EEHV4 genome compared to EEHV1 and HCMV. The dot matrix diagrams showing direct linear nucleotide alignments were generated as implemented at http://blast.ncbi.nlm.nih.gov/Blast.cgi. (a) Comparison across the intact 206-kb genome of EEHV4(Baylor) (KT832477) from the GC-rich branch of the Proboscivirus genus with the intact 180-kb genome of EEHV1A(Kimba) (KC618527) from the AT-rich branch of the Proboscivirus genus derived from the work of Ling et al. (24) when aligned in the same orientation. (b) Comparison across the intact 206-kb genome of EEHV4(Baylor) (KT832477) with the intact 235-kb genome of HCMV(Merlin) (AY446834.2) in the Cytomegalovirus genus of the mammalian betaherpesvirus subfamily derived from the work of Dolan et al. (41), with the latter aligned in the standard orientation.