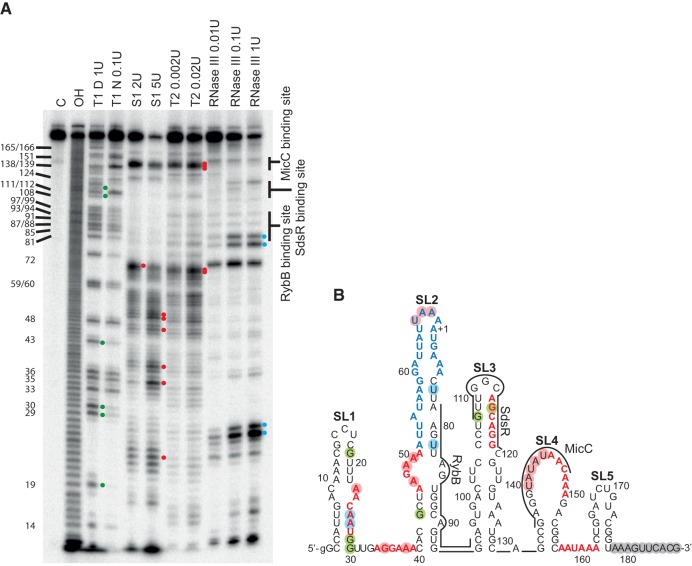
FIGURE 4.
The binding sites of RybB, SdsR, and MicC sRNAs have different structural contexts within the ompD mRNA leader. (A) The structure probing of 5′-32P-labeled ompD-187 using RNases indicated above the lanes. The untreated ompD-187 sample was resolved in lane C, formamide ladder in lane OH, and the products of reactions with RNase T1 in denaturing or native conditions in lanes marked T1 D and T1 N, respectively. The positions of G-specific cleavages by RNase T1 are indicated on the left side of the gel. (B) The secondary structure model of ompD-187 proposed by the RNAstructure program with hard constraints (circled positions) obtained from structure probing experiments in A. Residues constrained as single-stranded are indicated with red circles, while double-stranded are indicated with blue circles (RNase III) or green circles (RNase T1 in native conditions). Gray circles indicate nucleotides constrained as single-stranded based on experiments comparing the cleavage patterns of ompD-187 and 3′-end-truncated ompD-177 (data not shown). The AU-rich region is marked in blue, and (ARN)2 motifs in red. The binding sites of RybB, SdsR, and MicC sRNAs are indicated by lines.