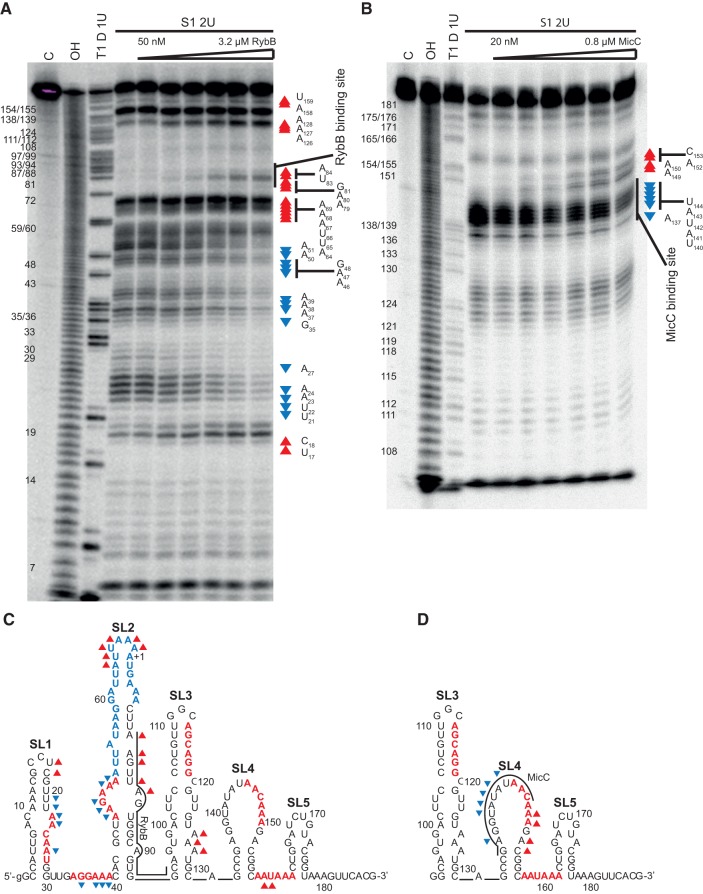
FIGURE 5.
The binding of RybB and MicC sRNAs induces local conformational changes in the ompD mRNA leader. (A) The structure probing of 5′-32P-labeled ompD-187 with nuclease S1 at increasing concentrations of RybB sRNA. (B) The structure probing of 5′-32P-labeled ompD-93–187 with nuclease S1 at increasing concentrations of MicC sRNA. The untreated samples were resolved in lanes marked C, formamide ladders in lanes OH, and reactions with RNase T1 in denaturing conditions in lanes marked T1 D. The positions of G-specific cleavages by RNase T1 are indicated on the left sides of the gels. The changes in nucleotide susceptibility to cleavage upon RybB or MicC binding are shown on the secondary structure of ompD-187 in C, and ompD-93-187 in D, respectively. The nucleotide positions, which showed higher susceptibility to degradation by nuclease S1 in the presence of respective sRNA, are marked by red triangles, and those which showed lower susceptibility are marked by blue reverse triangles.