Abstract
The fungal cell wall not only plays a critical role in maintaining cellular integrity, but also forms the interface between fungi and their environment. The composition of the cell wall can therefore influence the interactions of fungi with their physical and biological environments. Chitin, one of the main polysaccharide components of the wall, can be chemically modified by deacetylation. This reaction is catalyzed by a family of enzymes known as chitin deacetylases (CDAs), and results in the formation of chitosan, a polymer of β1,4-glucosamine. Chitosan has previously been shown to accumulate in the cell wall of infection structures in phytopathogenic fungi. Here, it has long been hypothesized to act as a 'stealth' molecule, necessary for full pathogenesis. In this study, we used the crop pathogen and model organism Magnaporthe oryzae to test this hypothesis. We first confirmed that chitosan localizes to the germ tube and appressorium, then deleted CDA genes on the basis of their elevated transcript levels during appressorium differentiation. Germlings of the deletion strains showed loss of chitin deacetylation, and were compromised in their ability to adhere and form appressoria on artificial hydrophobic surfaces. Surprisingly, the addition of exogenous chitosan fully restored germling adhesion and appressorium development. Despite the lack of appressorium development on artificial surfaces, pathogenicity was unaffected in the mutant strains. Further analyses demonstrated that cuticular waxes are sufficient to over-ride the requirement for chitosan during appressorium development on the plant surface. Thus, chitosan does not have a role as a 'stealth' molecule, but instead mediates the adhesion of germlings to surfaces, thereby allowing the perception of the physical stimuli necessary to promote appressorium development. This study thus reveals a novel role for chitosan in phytopathogenic fungi, and gives further insight into the mechanisms governing appressorium development in M.oryzae.
Author Summary
Magnaporthe oryzae is a filamentous fungal pathogen which causes devastating crop losses in rice. Successful invasion of the host is dependent upon the ability of the fungus to remain undetected by the innate immune system of the plant, which recognizes conserved components of the fungal cell wall, such as chitin. Previous studies have demonstrated that infection-related changes in cell wall composition are necessary to allow the fungus to remain undetected during infection. One such change that has long been hypothesized to have a role as a 'stealth mechanism' is the deacetylation of the polysaccharide chitin by enzymes known as chitin deacetylases. The deacetylation of chitin produces a polysaccharide known as chitosan, which has previously been shown to accumulate specifically on infection structures in plant pathogenic fungi. However, in this study, we show that germling-localized chitosan is not required for pathogenicity, arguing against a role as a 'stealth mechanism' at this stage. Instead, chitosan is required for the development of the appressorium, a critical fungal infection structure required for the penetration of plant cells. This requirement can be attributed to chitosan mediating the adhesion of germlings to surfaces, which is required for the perception of physical stimuli.
Introduction
All fungal cells are encased within a cell wall. This complex and dynamic structural barrier is composed of interwoven polysaccharides and proteins. Indeed, the polysaccharide moiety makes up the majority of the fungal wall, being comprised of chitin (a polymer of β1,4-N-acetylglucosamine), β/α1,3-glucans and mannans [1]. There is, however, considerable variation in the proportion of these wall components between different cell-types and between fungal species [1]. The cell wall plays an essential role in maintaining cellular integrity in the face of the challenging and varied environmental conditions to which fungi are exposed. Indeed, the cell wall plays a far greater role than simply being the extracellular “coat of armour” [2]. Firstly, morphogenesis is heavily influenced by cell wall composition, and, conversely, localized changes in composition allow fungal cells to undergo morphogenesis, whilst maintaining cellular integrity [3]. This is illustrated by many studies, where chemical or genetic perturbation of cell wall composition has led to gross defects in fungal growth and morphogenesis (reviewed in [1, 4]). Secondly, the cell wall forms the interface between the fungal cell and its immediate environment. As a result, the structure and composition of the cell wall influences both physical and biological interactions which occur over the life-cycle of the fungus, as, for example, during host infection [2].
The hemibiotrophic fungus Magnaporthe oryzae causes significant losses of rice [5]. It is thus a notable pathogen but is also considered as a model organism to study appressorium formation in pathogenic fungi [6]. Disease occurs when three–celled asexual conidia land on the host, differentiate a short germ tube and thence an infection structure (the appressorium). This developmental progression occurs in response to hard, hydrophobic surface and following the perception of host-derived surface chemistries, such as cutin [7]. The maturing appressorium generates a considerable turgor pressure, a penetration peg emerges and penetrates the leaf cuticle [8]. Subsequently, invasive hyphae ramify though the host. Throughout this process the fungal wall undergoes extensive remodelling during rapid growth [9].
For successful infection M. oryzae must remain undetected by its host: but how does the fungus do this? Plants readily detect key molecular signatures of fungal cells, that is, Pathogen Associated Molecular Patterns (PAMPs). Fungal cell wall polysaccharides in particular represent a major source of PAMPs [10]. The best characterized of these are chitin oligomers, which are released from the fungal wall either as a result of endogenous cell wall remodelling, or due to the action of plant chitinases. Chitin oligomers are recognized in rice plants by the Pattern Recognition Receptor (PRR) CEBiP (Chitin Elicitor Binding Protein) [11]. Binding of chitin oligomers to CEBiP induces its dimerization and, in association with CERK1 (Chitin Elicitor Receptor Kinase-1), results in phosphorylation and activation of CEBiP [12]. This triggers PAMP Triggered Immunity (PTI) [13]. This response includes callose deposition, the production of degradative enzymes (chitinases and glucanases), and a burst of reactive oxygen species [14, 15]—all of which serve to restrict or kill the invading fungus. Phytopathogenic fungi have evolved a number of strategies to avoid triggering PTI. The first is by secretion of chitin-binding effectors, which compete with the chitin receptors to bind the oligomers, and thus enable full pathogenesis, as in Cladosporium fulvum and M.oryzae [16, 17]. The second strategy is to change cell wall composition to either alter identity, or to mask the presence of the PAMPs. For example, during infection-related development, M.oryzae synthesises α1,3-glucan as a component of the cell walls of germ tubes, appressoria and invasive hyphae [9]. Deletion of the sole gene encoding α1,3-glucan synthase (AGS1) results in enhanced susceptibility to enzymatic degradation, the triggering of PTI and thus loss of pathogenicity [18]. This suggests that α1,3-glucan acts as a 'stealth' mechanism, cloaking the fungus from recognition by the plant. We postulated that chitin deacetylation may play a similar role—as it constitutes another infection-related cell wall alteration, observed to occur in a number of fungal plant pathogens, including Uromyces fabae, Colletotrichum graminicola, Puccinia graminis and M.oryzae [9, 19].
Chitin deacetylation is catalyzed by a family of conserved carbohydrate esterase enzymes (CE-4 family) known as chitin deacetylases (CDAs) (E.C. 3.5.1.41). The removal of acetyl groups from N-acetylglucosamine residues of chitin results in the formation of chitosan, typically a heterogeneous polymer of β1,4-glucosamine and β1,4-N-acetylglucosamine [20]. The deacetylation of chitin confers two hypothetical benefits upon the invading fungus. Firstly, unlike chitin, chitosan cannot cause activation of CEBiP [12]. In addition, although various defence responses have been observed in plants treated with chitosan (reviewed in [21], it is unclear whether chitosan is as effective as chitin in this respect. Secondly, chitin deacetylation likely provides protection from plant chitinases, since chitosan is a poor substrate for these enzymes [22]. However, such roles for chitosan, acting as either a protectant or as a 'stealth' mechanism are not yet supported by direct experimental evidence. Moreover, chitin deacetylases genes are present in the genomes of all fungi [23]. Thus, chitosan may be a broadly-occurring cell wall component, with roles beyond those we hypothesize for plant pathogens. Indeed, such additional roles may relate to the different physical and chemical properties of chitin and chitosan. Chitin occurs in fungal cell walls as antiparallel chains, forming microfibrils with immense strength and rigidity [24]. However, chitin deacetylation creates primary amine groups, which are largely protonated and charged at physiological pH. Consequently, chitosan is polycationic, more hydrophilic and an amorphous polysaccharide, in stark contrast to its parent polymer chitin. Thus, deacetylation could have profound consequences on the structure and physical properties of the fungal cell wall, which, in turn, will impact on fungal growth and morphogenesis.
There is, however, little published data defining the role(s) of chitosan in fungal cell walls per se. The role of chitin deacetylation was first described in S.cerevisiae [25] Here, two functionally redundant CDAs deacetylate chitin specifically in the ascospore cell wall—the double knockout mutant Δcda1Δcda2 strain results in complete loss of chitosan from the ascospore wall. Whilst spore viability was unaffected, spores of the Δcda1Δcda2 strain were more susceptible to treatment with lytic enzymes, ether and heat shock, suggesting possible disorganization and increased permeability of the wall [25, 26]. Further experimentation showed that the outer dityrosine layer was absent in CDA deletion strains [27]. It is hypothesized that the dityrosine is cross-linked to the amine groups of the chitosan, thereby creating a rigid and impermeable ascospore cell wall [28]. Similarly, chitosan was shown to be a component of spore wall in Ashbya gossypii—deletion of the single CDA resulted in a complete loss of sporulation, suggesting that chitosan is required for spore development [29]. In the Basidiomycete fungus Cryptococcus neoformans, chitosan is a major component of the vegetative cell wall [30]. Deletion of 3 of the 4 CDA genes (CDA1-3) abolished chitosan synthesis [31]—these triple deletion strains were hypersusceptible to various cell wall perturbants, showed attenuated virulence and “leaked” melanin from their cell walls [32] -Chitosan is thus required for cell wall integrity in this fungus..
Deacetylation of chitin may play critical roles in fungal development, integrity and pathogenesis. Thus the two central hypotheses regarding its role are: i) chitin deacetylation is important for cellular development and morphogenesis and ii) chitosan acts mechanisitically as a 'stealth molecule' during plant infection in pathogenic fungi. Appressorium development in M.oryzae represents an amenable and relevant system to test these hypotheses. Firstly, chitosan is known to be a component of the germ tube and appressorium cell wall [9]. Secondly, protective cell wall components (α1,3-glucan) in appressoria are required for pathogenicity [18]; this may hold true for chitosan. Lastly, appressorium development likely requires increased cell wall flexibility—chitin deacetylation may enable such plasticity. Indeed, this may be the most likely scenario as deletion of a putative chitin deacetylase, CBP1 resulted in defective appressorium formation on artificial surfaces [33]. Yet, deletion of CBP1 also prevented hyphopodium and pre-invasive hypha formation on artificial surfaces (but not root surfaces), suggesting that chitosan plays a role in multiple infection-related cellular differentiation events [34]. However, chitosan synthesis was not consistently reduced in the cbp1 mutant [34] and neither was the chitin deacetylase activity of Cbp1 proven [33] and so the role of chitin deacetylation remains inconclusive. A comprehensive characterization of chitin deacetylases operating during appressorium development should provide a valuable insight into the roles of chitin deacetylation during infection in plant pathogenic fungi.
Results
Chitin deacetylation occurs during appressorium development
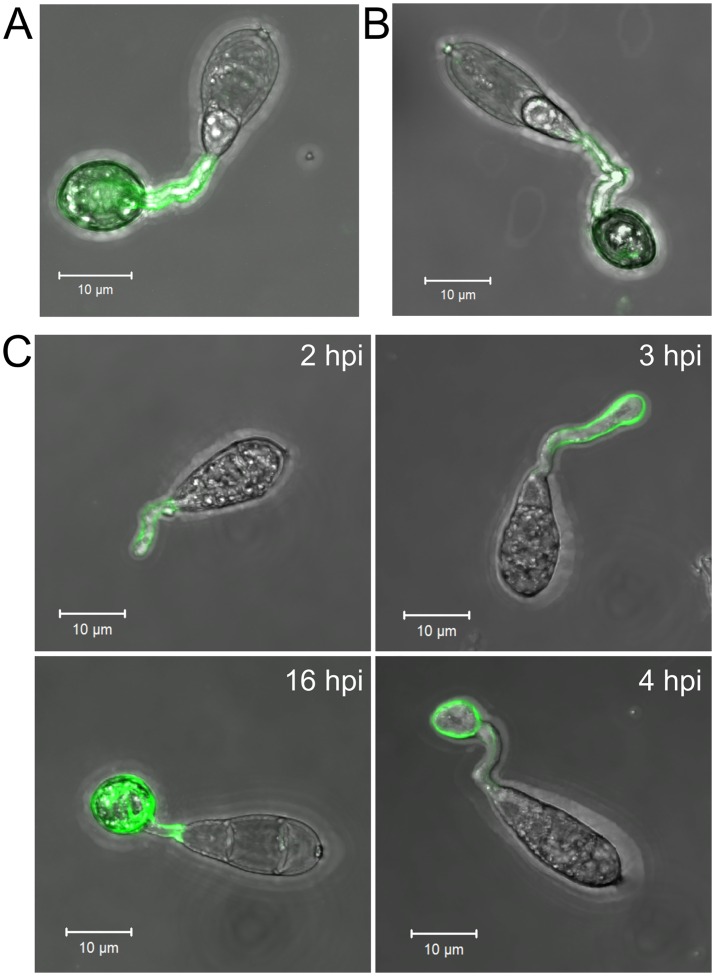
The localization of chitosan during appressorium development in M.oryzae has previously been investigated using Eosin Y (9). Whilst this dye binds to chitosan, via an electrostatic interaction, the specificity of this interaction has not been directly proven. We sought an alternative, more specific method, that is by immune-detection, with mAbG7, a monoclonal anti-chitosan antibody [35], and a polyclonal anti-chitosan antibody [19]. These antibodies revealed that chitosan localizes to the cell wall of the germ tube and the appressorium (Fig 1A & 1B), and is consistent with previous observations made with Eosin Y [9]. However, we wished to determine the precise timing of chitin deacetylation during appressorium development in order to assess whether it is, indeed, required for this process. To do this, we invoked use of the probe OGA488. This recently developed and highly specific probe [36] stains chitosan rapidly (within 15 minutes), and is thus ideally-suited to studying germlings in vivo at different stages of morphogenesis. Using this probe, germling development was tracked on an artificial hydrophobic surface inductive to appressorium formation [37]. After 1–2 hpi (hours post inoculation) the germ tube was weakly labelled, labelling strengthened upon onset of germ tube hooking (2–3 hpi), and intensified further during appressorium development (4 hpi). At this stage, the entire germ tube and appressorium wall were labelled—this being consistent with the immune-localisation data, (Fig 1C).
Fig 1. Chitosan is a component of the cell wall in germ tubes and appressoria.
Germlings, labelled with A) monoclonal anti-chitosan antibody mAbG7, B) polyclonal anti-chitosan antibody (staining with antibodies was performed at 16 hpi) and C) The anti-chitosan probe OGA488 which localized chitosan in live cells at different stages of appressorium development (2–16 hpi, as indicated). Germlings were inoculated onto a hydrophobic glass surface (inductive to appressorium formation). Scale bars: 10 μm.
Chitin deacetylases are expressed during appressorium development
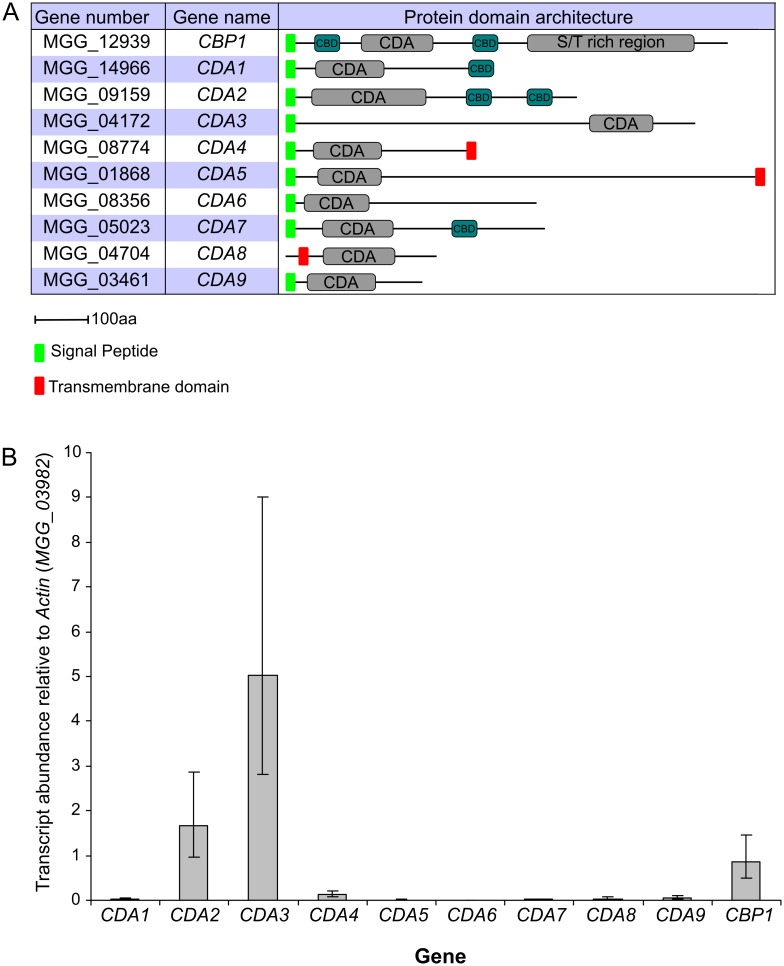
Next, we asked which of the 10 putative chitin deacetylase genes residing in the M.oryzae genome (http://www.broadinstitute.org/annotation/genome/magnaporthecomparative/MultiHome.html) are responsible for chitin deacetylation occuring prior to and during appressorium development. The 10 M.oryzae CDAs carry a polysaccharide/chitin deacetylase domain (Pfam 01522). All, except Cda9, have a predicted N-terminal signal peptide. Putative chitin binding domains (CBD; Pfam 00187) are present in Cbp1, Cda1, Cda2 and Cda7. Cbp1 also has a serine/threonine-rich repeat region at its C-terminus [33]. Cda4, Cda5 and Cda8 have putative single transmembrane domains, suggesting that these proteins localise to the membrane (Fig 2A).
Fig 2. M.oryzae has 10 putative chitin deacetylases.
A) Table of the 10 putative chitin deacetylases in M.oryzae, showing gene numbers, given gene names and protein domain architectures (as defined by Pfam). B) Expression of the chitin deacetylases during appressorium development on rice leaves. Leaves of the susceptible cultivar CO-39 were inoculated with conidia of M.oryzae, and incubated for 5 hr, to induce appressorium development. Expression levels of the CDAs were analyzed by qRT-PCR and normalized to Actin (MGG_03982). Error bars show standard deviation, n = 3.
Chitin deacetylation in Colletotrichum lindemuthianum is dependant upon a zinc-binding Asp-His-His triad, and four key active site residues: a catalytic base residue (Asp) and a catalytic acid residue (His), which interact with Arg and Asp residues respectively [38, 39]. Sequence alignment using Clustal Omega [40], revealed conservation of the zinc-binding triad and active-site resides in all M.oryzae CDA sequences, with the exception of a non-catalytic Asp in Cbp1 (MGG_12939) at position 153 of the alignment (S1 Fig).
To determine which of the 10 CDAs are expressed during appressorium development, we performed qRT-PCR analysis. Total RNA was extracted from rice leaves inoculated with WT conidia, at 5 hpi. Appressorium development was confirmed at this timepoint by microscopic observation (S2 Fig). Relative quantification of transcript abundances for all 10 CDAs revealed that CDA3 is the most highly expressed, followed by CDA2 and CBP1 (Fig 2B). Low levels of expression were observed for the remaining genes. In addition to this, we also analyzed the expression of the CDAs during later stages of leaf infection, at 36 hpi. In this case, CDA1 and CDA6 showed the highest transcript abundance (S2 Fig). These data align with published transcriptomics datasets [41, 42].
Chitin deacetylases are required for appressorium development on artificial surfaces
The combined transcriptional profiling data reveals that three particular CDA genes are likely involved in chitin deacetylation during appressorium development: CBP1, CDA2, and CDA3. CBP1 has previously been partially characterized [33], but we wished to further this work, in combination with CDA2, and CDA3 and thus generated single, double and triple deletion strains by a targeted gene replacement approach. All such mutants generated were confirmed by both PCR and by Southern Blot analysis (S3 and S4 Figs). Deletion strains are listed in S1 Table.
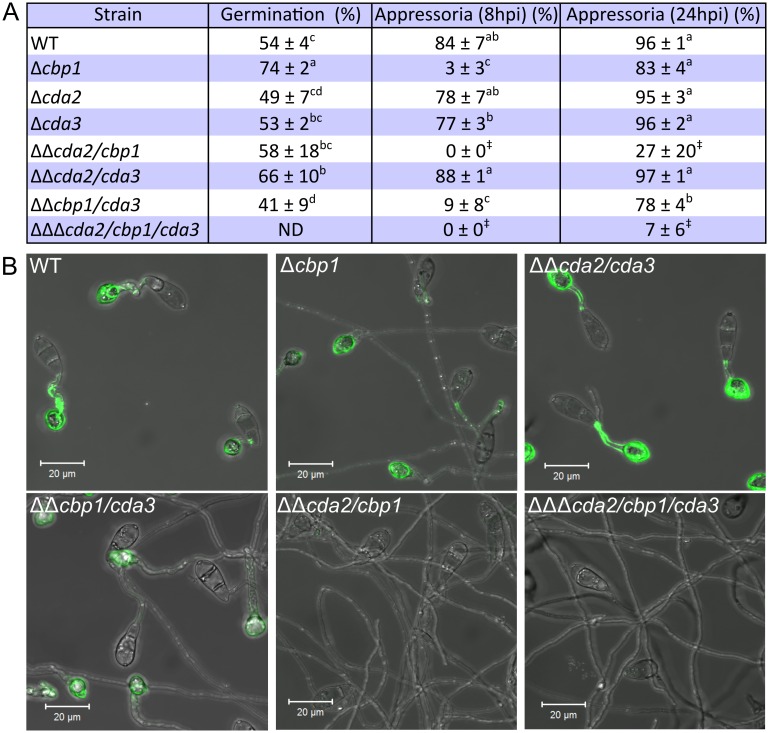
To compare appressorium development, conidia of the mutant and WT strains were inoculated onto an artificial hydrophobic glass surface. In the WT strain, over 80% of conidia developed appressoria after an 8 hr incubation (Fig 3A). However, very few appressoria formed in cbp1 at 8 hpi (3% ± 3% (SD n = 3))—germ tubes were abnormally elongated and failed to hook at the end distal from the spore (Fig 3B). This is consistent with the previous report [33]. Germination of cbp1 conidia was significantly higher than the WT strain at 1 hpi (Welch's ANOVA, p < 0.05, Fig 3A) and thus failure to form appressoria is not due to delayed or reduced germination. By 24 hpi, 83% ± 4% of cbp1 germ tubes formed appressoria (Fig 3A) that were misshapen in appearance (S6 Fig). Appressorium development is therefore both delayed and defective in cbp1. In the cda2, cda3 and cda2/cda3 strains, appressorium development and conidial germination progressed similarly to WT. However, deletion of CDA2 together with CBP1 resulted in much more severe defects in appressorium development than observed in the single deletion strain cbp1. Germlings of strain cda2/cbp1 failed to form appressoria by 8 hpi, as in cbp1, but extending the incubation period to 24 hpi resulted in only rare occurrences of appressorium development (27% ± 20 of germ tubes), in contrast to cbp1 (Fig 3A). The germ tubes of cda2/cbp1 appeared even more elongated (487 ± 42 μm in cda2/cbp1 compared with 264 ± 37 μm in cbp1 (S3 Table)), yet remained undifferentiated. This phenotype was also observed when conidia were germinated on other hydrophobic surfaces, including plastic and Parafilm (S5 Fig). CDA2 and CBP1 therefore exhibit partial redundancy. Conversely, deletion of CDA3 had no additive effects on appressorium development under the conditions tested: cbp1/cda3 demonstrated an identical phenotype to cbp1, and cda2/cbp1/cda3 a similar phenotype to cda2/cbp1.
Fig 3. Appressorium development and chitin deacetylation are dramatically reduced in the CDA deletion strains.
A) Table showing percentage conidial germination (at 1 hpi) and percentage of germ tubes forming appressoria (at 8 and 24 hpi) on a hydrophobic glass surface (± SD, n = 3). Germination rates were analyzed using Welch's ANOVA, with Games-Howell post-hoc test. Means not sharing a letter indicate significant differences (p < 0.05). Significantly reduced appressorium formation frequency was observed in certain deletion strains (1-way ANOVA with post-hoc Tukey test. Means not sharing a letter indicate significant differences p < 0.05). ‡: data could not be analyzed using ANOVA, as did not fulfil requirements. B) Fluorescent labelling of chitosan on germlings of the CDA deletion strains at 16 hpi, showing complete lack of staining in the cda2/cbp1 and cda2/cbp1/cda3 strains. Scale bars: 20 μm.
To determine whether chitin deacetylation is reduced in the cda mutants, germlings of the deletion strains were stained with OGA488 (Fig 3B). WT germlings developed on an artificial hydrophobic surface showed strong staining of the cell wall in germ tubes and appressoria at 16 hpi, as described previously. Similar staining was observed in cda2/cda3. In cbp1 and cbp1/cda3, appressoria labelled with OGA488, but little fluorescence was observed on their elongate germ tubes. However, no chitosan staining was observed in strains cda2/cbp1 and cda2/cbp1/cda3, suggesting complete loss of chitin deacetylation activity, and providing further evidence of functional redundancy between cbp1 and cda2. By 24 hpi a small proportion of germ tubes in strains cda2/cbp1 and cda2/cbp1/cda3 formed appressoria. However, no chitosan was detected (S6A Fig), suggesting that it may not be an absolute requirement for appressorium morphogenesis. Yet, the presence of low concentrations of chitosan, or chitosan in cell wall regions inaccessible to the OGA488 probe cannot be completely discounted.
The highly elongate germ tubes observed in strains cbp1, cda2/cbp1 and cda2/cbp1/cda3 are a curious feature. Calcofluor White staining revealed that the mutant germ tubes are septated, with cross wall distributed along the elongate germ tubes of cbp1, cda2/cbp1 and cda2/cbp1/cda3 at regular intervals (S6B Fig). Taken together, these data suggest a clear link between the loss of chitin deacetylation, and loss of appressorium development.
Localization of chitin deacetylases
We created fluorescent protein fusions, to better understand how chitin deacetylation by Cbp1 and Cda2 promotes appressorium development. C-terminal mCherry fusions of CBP1 and CDA2 were made, under the control of their respective native promoters, and transformed into their respective deletion background strains. Several independent transformant lines were characterized for each fusion and which exhibited identical patterns of fluorescence (S1 Table).
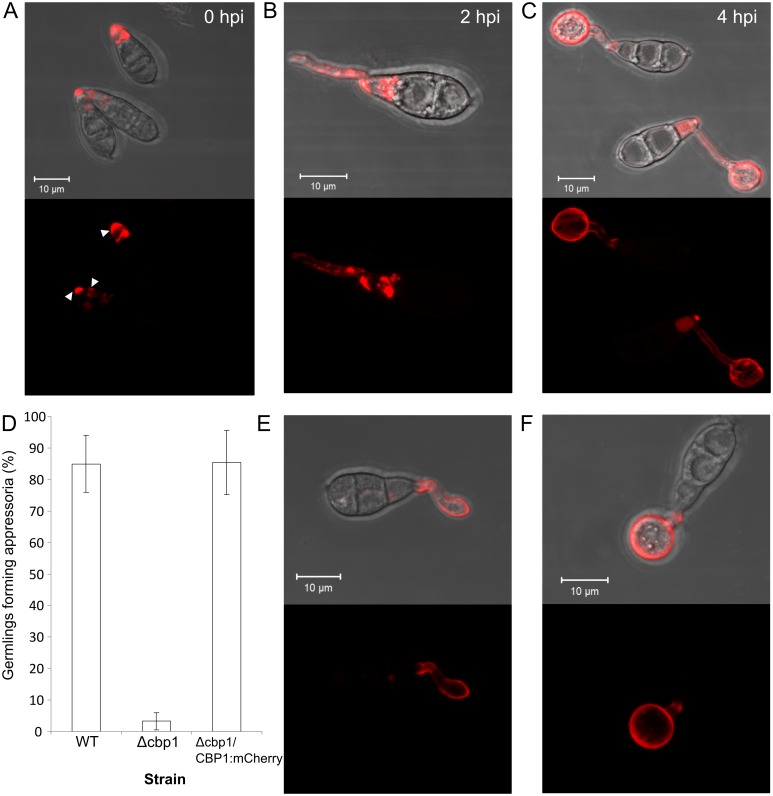
Strains expressing Cbp1:mCherry show fluorescence at all stages of germling morphogenesis (Fig 4A–4C), and demonstrate fully restored appressorium development (Fig 4D), indicating that the Cbp1:mCherry fusion protein is fully functional. Unexpectedly, fluorescence was observed in the cell wall at conidial apices, even in conidia which had not yet been harvested from the mycelium (Fig 4A). Substantial intracellular fluorescence was also observed at this stage, most likely localized to vacuoles. During initial stages of germ tube growth (1–2 hpi), weak fluorescence was observed at the lateral walls of the germ tube, but not the tip (Fig 4B). In addition, small punctae of intracellular fluorescence were sometimes observed along the length of the germ tube. At later stages of appressorium development (3–5 hpi), wall-localized fluorescence became stronger and localized to the entire germ tube and appressorium, although intracellular fluorescence was still apparent in some appressoria (Fig 4C). The appressorial wall remained fluorescent even at later stages (8–16 hpi, S7 Fig), although intensity decreased noticeably.
Fig 4. Localization of mCherry tagged Cbp1 and Cda2.
Confocal fluorescence microscopy images of germlings expressing Cbp1:mCherry, showing fluorescence in conidia, germ tubes and appressoria at 0, 2 and 4hpi on hydrophobic glass (A-C). D) Successful complementation of CBP1:mCherry fusion construct. Appressorium development was fully restored in the complemented strain at 8 hpi. Error bars show SD n = 3. E & F) Confocal fluorescence microscopy images of germlings expressing Cda2:mCherry at 2 hpi (E) and 4 hpi (F). Scale bars: 10 μm.
Similarly to Cbp1:mCherry, strains expressing Cda2:mCherry exhibited fluorescence during appressorium development, but with fluorescence typically appearing after hooking of the germ tube (Fig 4E). Strong fluorescence was also observed during subsequent appressorium formation (Fig 4F), and remained present at later stages (8 hpi), as observed with Cbp1:mCherry (S7 Fig). Unlike Cbp1:mCherry however, fluorescence was only very rarely observed intracellularly. Since the cda2 strain was apparently identical to the WT strain, functionality of the fusion protein could not be determined in the complemented strain. Attempts to create a Cda3:eGFP fusion protein did not yield fluorescent transformants, suggesting that the fusion protein is unstable.
Exogenous chitosan can rescue cda mutants
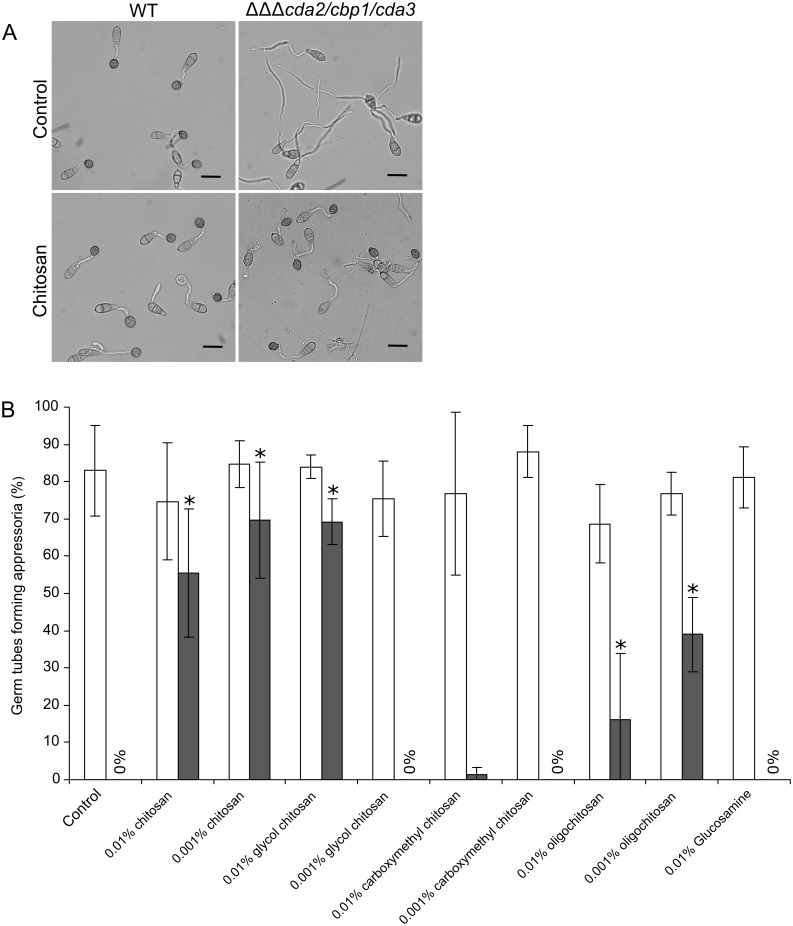
The defects in appressorium development observed upon deletion of CDA genes was similar to that resulting from deletion of the hydrophobin gene MPG1 [43]. It was reported that appressorium development could be rescued in the mpg1 mutant by co-inoculation with WT conidia, suggesting that Mpg1 could act in trans [44]. To test whether chitosan could rescue the cda mutants, exogenous chitosan was added to conidia during germination on an artificial hydrophobic surface. Addition of 0.01% (w/v) or 0.001% (w/v) chitosan, surprisingly, restored appressorium development in all cda deletion strains (Fig 5A & 5B). Between 70–85% of cbp1, cda2/cbp1 and cbp1/cda3 conidia formed appressoria after an 8hr incubation in the presence of 0.01% or 0.001% chitosan, compared with 0–10% in the control treatment (water). In the triple deletion strain (cda2/cbp1/cda3) rescue was slightly lower (55% ± 17 (SD, n = 3) and 70% ± 16 (SD, n = 3) appressoria at 8 hpi for 0.01% and 0.001% chitosan, respectively. In the WT strain, 70–85% of conidia formed appressoria in all treatments. To further investigate this, different derivatives and formulations of chitosan were tested for their ability to restore appressorium development in the deletion strains (structures are shown in S8 Fig). Glycol chitosan, a soluble derivative of chitosan restored appressorium development in cda2/cbp1/cda3 at a concentration of 0.01% but not 0.001%. Carboxymethylchitosan (in which chitosan has been modified by the addition of an anionic carboxymethyl group) did not restore appressorium formation at either concentration. Lastly, the addition of chitosan in oligomeric form (oligochitosan, with a degree of polymerization of 22–33 residues) partially restored appressorium formation at 8 hpi, whereas glucosamine showed no restorative effects at all. Taken together, these data suggest that the cationic nature of exogenous chitosan is the most important factor determining rescue of the mutant phenotype, with steric factors also having some influence. Polymer length appears to be of little importance, given the broad ranges capable of restoring appressorium development.
Fig 5. Exogenous chitosan can rescue appressorium development in CDA deletion strains.
A) Rescue of appressorium development in the cda2/cbp1/cda3 deletion strain with exogenously applied chitosan (0.01% w/v). Pictures taken at 8 hpi, scale bars: 20 μm. B) Comparison of appressorium development in WT (white bars) and cda2/cbp1/cda3 deletion strain (grey bars) in the presence of different derivatives and formulations of chitosan (shown in S8 Fig). Conidia were germinated on an artificial inductive surface for 8 hr in the presence of different types of chitosan at a final concentration of 0.01% or 0.001% (w/v), and mature appressoria were counted. Significant increases in appressorium development were observed in the cda2/cbp1/cda3 deletion strain (Asterisks, Student's T-test, p < 0.05). Error bars show SD, n = 3. DDA: degree of deacetylation.
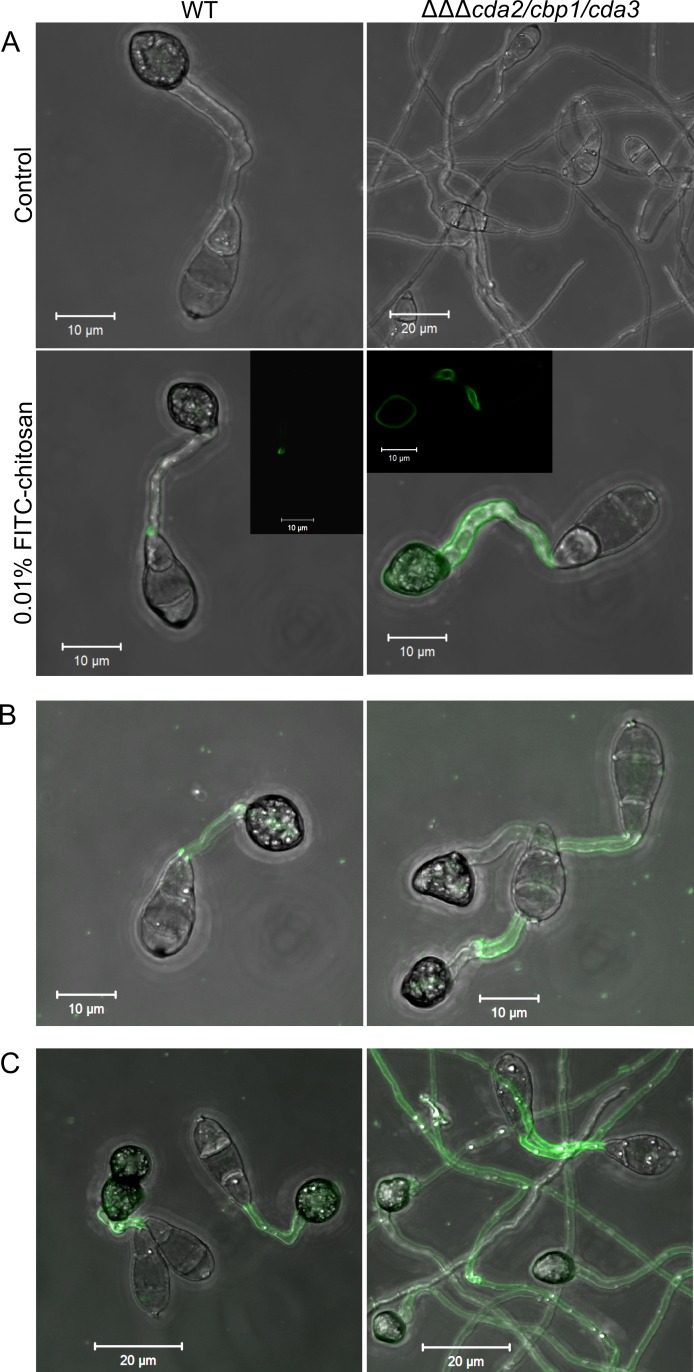
To investigate how exogenous chitosan restores appressorium development in the cda mutants, fluorescently-labelled chitosan was used. FITC-chitosan, synthesized according to Qaqish et al [45], was added to conidia of the WT and cda2/cbp1/cda3 strains on an artificial hydrophobic surface. After 16 hr incubation, the FITC-chitosan was removed and the germlings imaged by confocal microscopy. The FITC-chitosan restored appressorium development in cda2/cbp1/cda3, and clear localization to the cell wall of germ tubes and appressoria (Fig 6A). In the WT strain, weak fluorescence was occasionally observed in the germ tubes. Exogenous chitosan is therefore associated with the cell wall of the deletion strains.
Fig 6. Fluorescently labelled chitosan is associated with the cell wall of the CDA deletion strains.
A) Rescue of appressorium development in the cda2/cbp1/cda3 deletion strain using exogenous, fluorescently-labelled chitosan, following a 16 hr incubation, showing clear localization to the cell wall of the germ tube and appressorium (single z-section insert). B) Rescue of appressorium development after a 2 hr incubation with FITC-chitosan, followed by washing and a further 16 hr incubation, showing germ tube-localized FITC-chitosan. C) Rescue of appressorium development after addition of FITC-chitosan at 16 hpi, and incubation for a further 8 hr. Scale bars: as shown in pictures.
Chitosan localizes to the germ tube prior to appressorium development (Fig 1C). To determine whether this germ tube-localized chitosan is sufficient to induce appressorium development, FITC-chitosan was added to germinating conidia of WT or cda2/cbp1/cda3 strains for 2 hr only, before being washed off, and the germlings left to develop for a further 16 hr. In this way, chitosan was only present during the initial germ tube stage of germling morphogenesis, and was removed before appressorium development occurred. Appressorium development was restored in the deletion strain by this short incubation with FITC-chitosan. Here, fluorescence was only observed in the germ tube, and not the appressoria of cda2/cbp1/cda3 germlings, with similar localization in the WT (Fig 6B). Thus germ tube-localized chitosan seems sufficient to induce appressorium development.
Lastly, to investigate whether chitosan restores appressorium development at later stages of germling morphogenesis, conidia were incubated for 16 hr, FITC-chitosan added, and the conidia incubated for a further 8 hr. Again, appressorium development was restored in the cda2/cbp1/cda3 strain, although here the germ tubes were highly-elongate (Fig 6C). Fluorescence was observed along the entire length of these elongate germ tubes, but appeared more intense at regions proximal to its point of emergence from the conidium. In this case, WT germlings also exhibited weak fluorescence in both germ tubes and appressoria.
Pathogenicity of the cda mutants
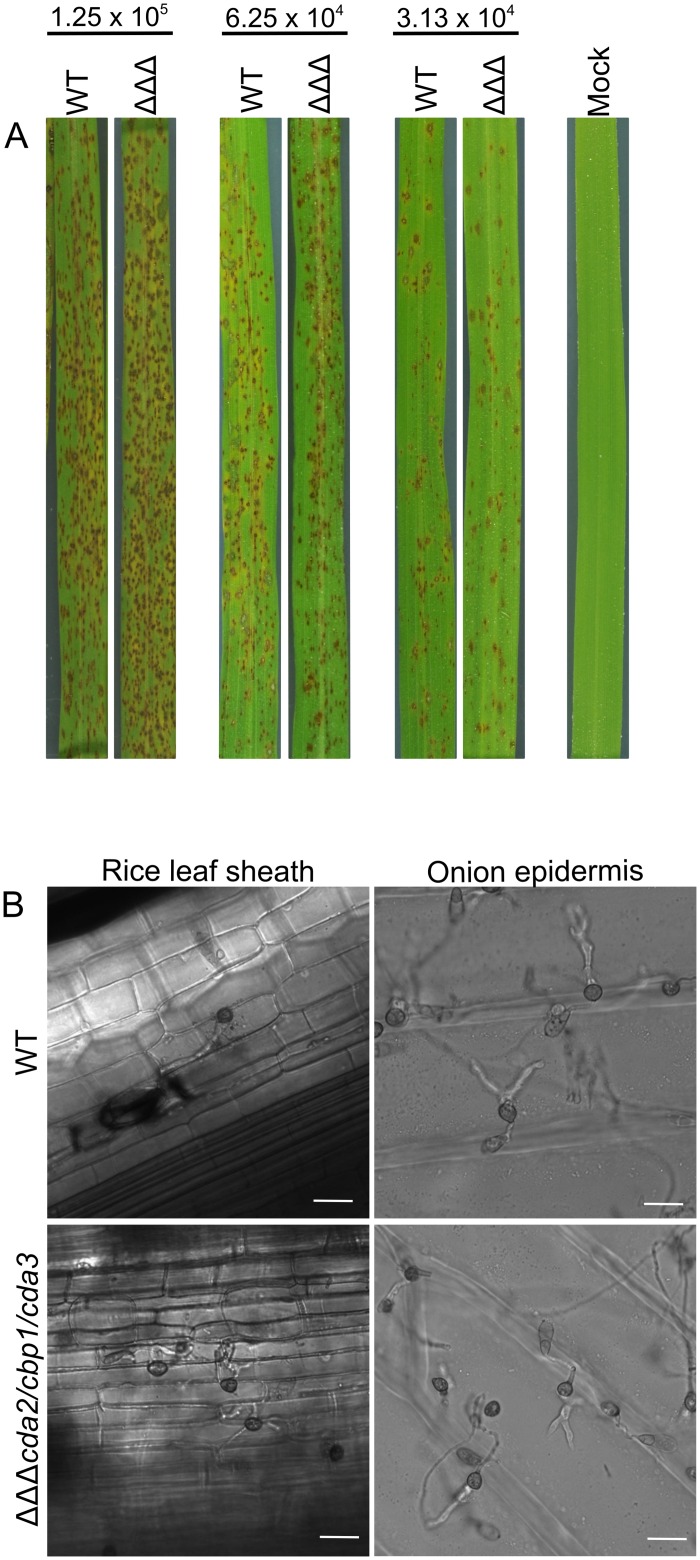
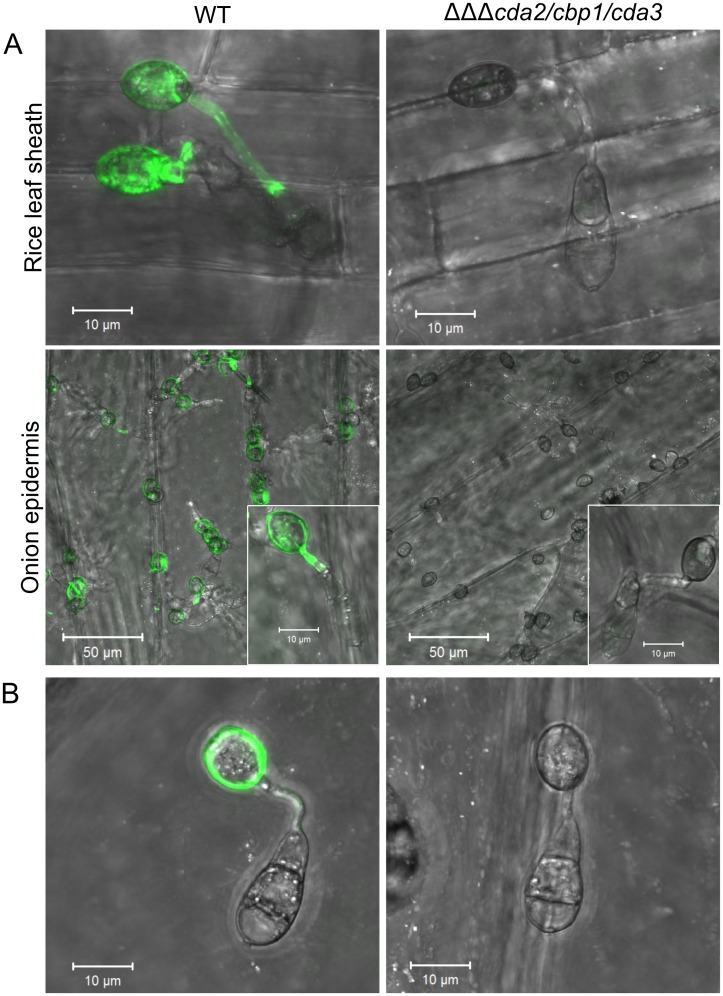
Appressorium development is highly defective in cda mutants on artificial hydrophobic surfaces, inductive to appressorium formation in the WT strain. To determine whether this results in a reduction in pathogenicity, equal concentrations of conidia of the deletion strains were inoculated onto detached rice leaves. As reported previously for cbp1 [33] and cda3 [46], and as expected for cda2 and cda2/cda3, pathogenicity was unaffected in these strains (S9 Fig). Surprisingly however, the double and triple deletion strains were also able to successfully infect detached rice leaves or whole plants, causing similar lesion numbers to the WT strain (Fig 7A and S9 Fig). Previously, appressorium development was found to be restored in the cbp1 strain on rice leaves [33]. To see if this was also the case in the other deletion strains, rice leaf sheaths were inoculated with conidia of the cda2/cbp1/cda3 strain, and incubated for 24 hr. This revealed that appressorium development was, indeed, restored upon germination on a leaf surface (Fig 7B). Not only this, but the appressoria of cda2/cbp1/cda3 were able to penetrate rice cells with similar efficiency to the WT: 67% (±7, (SD) n = 3) of WT appressoria had invasive hyphae at 24 hpi, compared with 47% (±10, (SD) n = 3) of cda2/cbp1/cda3 appressoria. To see if this effect was specific to the rice leaf surface, the experiment was repeated on onion epidermis. Again, appressorium development was restored in the cda2/cbp1/cda3 strain and invasive hyphae were observed at 24 hpi (Fig 7B).
Fig 7. Pathogenicity is unaffected in the cda mutants.
A) Pathogenicity of the WT and cda2/cbp1/cda3 strains on whole rice plants. Rice plants were inoculated with conidia of the WT or cda2/cbp1/cda3 strain at 3 different spore concentrations, and incubated for 4 days. The experiment was repeated 3 times, representative leaves are shown. B) Appressorium development in the WT and cda2/cbp1/cda3 strains on rice leaf sheath and on onion epidermis. Restored appressorium development and successful penetration of the plant cells was observed in cda2/cbp1/cda3. Scale bars: 20 μm.
Cuticular waxes can by-pass the requirement for chitosan
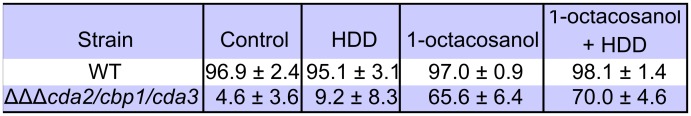
The artificial hydrophobic surface lacks the laminated layer of cutin sandwiched between wax on the rice leaf surface cuticular rice leaf [47]. To test whether wax is sufficient to restore appressorium development in the cda2/cbp1/cda3 strain, hydrophobic glass coverslips were coated with the wax molecule 1-octacosanol, which has previously been used to induce appressorium development in M.oryzae [48]. Conidia of strain cda2/cbp1/cda3 germinated on this surface for 24 hr demonstrated partially restored appressorium development (65.6 ± 6.4% on 1-octacosanol compared with 4.6 ± 3.6% on the control surface) (Fig 8). However, germ tubes of the triple deletion strain remained extremely elongated (S10 Fig).
Fig 8. Waxes can partially restore appressorium development in the cda2/cbp1/cda3 mutant.
Percentage of germ tubes demonstrating appressorium development in presence of 1-octacosanol and/or 1,16 hexadecanediol, comparing the WT and cda2/cbp1/cda3 strain at 24 hpi.
Cutin monomers are known to induce appressorium development in M.oryzae [7], To determine whether cutin restores appressorium development in the cda mutants, conidia of cda2/cbp1/cda3 were germinated on an artificial hydrophobic glass surface, in the presence of 1,16-hexadecanediol (HDD; cutin monomer) either on its own, or in combination with 1-octacosanol. HDD did not induce appressorium development in germlings of the cda2/cbp1/cda3 strain, and no synergistic effect was observed when HDD was used in combination with 1-octacosanol (Student's T-test, no significant difference at p < 0.05, compared with 1-octasosanol treatment alone) (Fig 8 and S10 Fig).
Chitosan is not required for appressorium development on plant surfaces
The rescue of appressorium development in the cda2/cbp1/cda3 strain on plant surfaces is perplexing. We hypothesized that chemical factors present on the leaf surface, such as wax, trigger upregulation of additional, functionally redundant chitin deacetylases, and restore chitin deacetylation. To test this, conidia of the cda2/cbp1/cda3 and WT strains were germinated on rice leaf sheaths or onion epidermis', and, subsequently stained with OGA488. Chitosan remained completely undetectable in the triple deletion strain, whilst strong labelling of germ tubes and appressoria was observed in the WT (Fig 9A). This suggests that restoration of appressorium development in cda2/cbp1/cda3 on plant surfaces is not due to the upregulation of redundant chitin deacetylases. Chitosan is thus not an absolute requirement for appressorium development; instead, this requirement appears to be surface dependant.
Fig 9. Restored pathogenic development on plant surfaces is not accompanied by restored chitin deacetylation.
Fluorescent labelling of chitosan with OGA488 in WT and cda2/cbp1/cda3 germlings germinated on A) rice leaf sheaths or onion epidermis for 24 hr, showing complete absence of chitosan in the cda2/cbp1/cda3 strain. B) Fluorescent labelling of chitosan in WT and cda2/cbp1/cda3 germlings germinated on onion epidermis for 4 hr, showing absence of chitosan in unmelanized appressoria of the deletion strain. Scale bars: as stated in pictures.
The appressorial cell wall is impregnated with an impermeable layer of melanin. This may mask detection of chitosan in the appressoria of cda2/cbp1/cda3. Mutant strain germlings were also stained with OGA488 at 4 hpi, prior to melanization of the appressoria. However, no OGA488 staining was detected in unmelanized cda2/cbp1/cda3 germlings, suggesting the melanin does not prevent the detection of chitosan (Fig 9B).
Chemical inducers can restore appressorium development in cda mutants
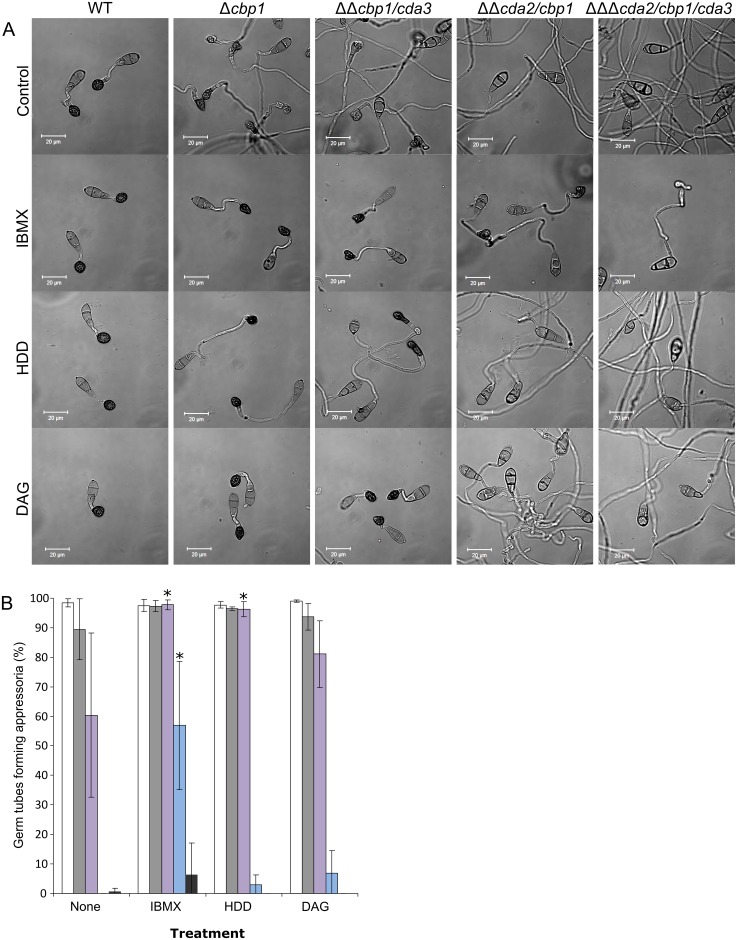
Appressorium development in M.oryzae is controlled by a number of regulatory pathways, which have been shown to respond to the physical and chemical cues present on the leaf surface [49]. In order to determine if there is interplay between chitosan and these particular relays, appressorium development assays were performed in the presence of chemical inducers of these pathways. Previously, appressorium development in cbp1 has been shown to be restored by the addition of cAMP, 1,16 hexadecanediol or diacylglycerol [33]. We tested the cda mutant strains by germinating conidia on a hydrophobic glass surface in the presence of 2.5 mM IBMX (a phosphodiesterase inhibitor which increases intracellular levels of cAMP), 200 mM 1,16 hexadecanediol (HDD, a cutin monomer) or 58 mM diacylglycerol (DAG, an activator of protein kinase C). Consistent with the previous report, all treatments restored appressorium development in cbp1 (Fig 10). Although the proportion of germ tubes forming appressoria was unchanged in this case (since observations were made at 16 hpi), germling morphology of cbp1 was now similar to WT, as evidenced by a significant reduction in germ tube lengths (Mann-Whitney U-test, p < 0.001 S3 Table). cbp1/cda3 showed a similar profile of sensitivity to the chemical inducers as cbp1. Intriguingly however, cda2/cbp1 was considerably less affected by all three chemical inducers: neither HDD or DAG caused a significant increase in appressorium development in this strain (Student's T-test, p < 0.05), whilst IBMX effected only a partial rescue towards the WT phenotype. In cda2/cbp1/cda3, IBMX treatment led to a small proportion of germ tubes forming aberrant appressoria by 16 hpi, although this increase was non-significant (Student's T-test, p < 0.05). Rescue of cbp1 by HDD and DAG is therefore dependent upon CDA2, and rescue by IBMX is dependent upon both CDA2 and CDA3.
Fig 10. The CDA deletion strains show distinct sensitivity to chemical inducers of appressorium development.
A) Micrographs of germlings at 16 hpi in presence or absence IBMX, 1,16 hexadecanediol (HDD), or diacylglycerol (DAG), showing drastically reduced inductive effects in cda2/cbp1 and cda2/cbp1/cda3 strains. Scale bars: 20 μm. B) Quantification of appressorium development, as visualized in A, WT: White bars, cbp1: grey bars, cbp1/cda: purple bars, cda2/cbp1: blue bars and cda2/cbp1/cda3: dark grey bars. Error bars show SD, n = 3. Significant differences in appressorium development were observed upon treatment with inducers in the cbp1/cda3 and cda2/cbp1 strains (asterisks, Student's T-test p < 0.05).
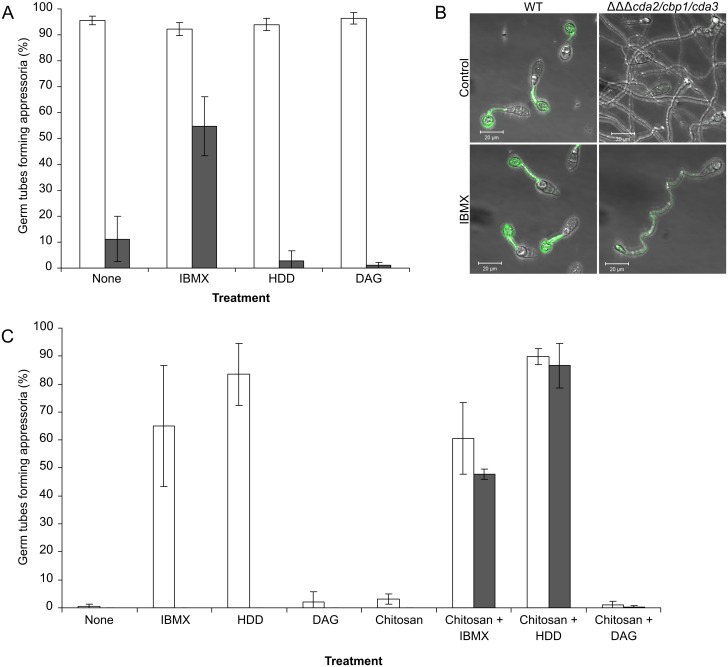
Hydrophobicity is one of the key physical cues inducing appressorium development, and is perceived in a cAMP-dependant manner [50]. In order to further examine the relationship between hydrophobic surface sensing, chitosan and the chemical inducers, germling differentiation assays, with the WT and mutant strain cda2/cbp1/cda3, were performed on both hydrophobic and a hydrophilic glass surfaces. At 24 hpi on the hydrophobic surface, the response of cda2/cbp1/cda3 germlings to the inducers was much the same as at 16 hpi, although the proportion of appressoria formed with IBMX was higher (Fig 11A). As with the appressoria formed on the plant leaf surface, it was important to determine whether this rescue could be explained by restored chitin deacetylation. IBMX-treated germlings of the WT and cda2/cbp1/cda3 strains were stained with OGA488 at 24 hpi. This revealed that chitosan was indeed present on the germ tubes and appressoria of the triple deletion strain, albeit to a much lesser extent than the WT germlings (Fig 11B). This suggests that rescue by IBMX may simply be due to upregulation of redundant chitin deacetylases, as observed in cbp1, rather than the activation of an otherwise inactive signalling pathway.
Fig 11. Combinations of chemical inducers are required to restore appressorium development in cda2/cbp1/cda3.
Induction of appressorium development in the cda2/cbp1/cda3 strain by chemical inducers, on A) hydrophobic and C) hydrophilic glass surfaces. Appressorium development could be partially restored in the triple deletion strain (grey bars) after a 24 hr incubation with IBMX, although chitosan staining of these germlings (B) showed that the appressorium development was associated with an increase in chitosan in the germlings of the cda2/cbp1/cda3 mutant. C) Appressorium development on hydrophilic surfaces could be restored to levels similar to the WT strain by combinations of 0.01% chitosan (w/v) and 2.5mM IBMX or with 200 μm 1,16 hexadecanediol (HDD). White bars: WT, grey bars: cda2/cbp1/cda3. Error bars: SD, n = 3.
Germinations were repeated on a hydrophilic glass surface (Fig 11C). No appressorium development was observed in the WT strain after a 24 hr incubation (consistent with previous observations [37]), but it was restored by the addition of IBMX or HDD. Both such inducers remained ineffective on cda2/cbp1/cda3. As exogenous chitosan fully restores appressorium development in the CDA deletion strains on a hydrophobic surface we extended this work to evaluate its effect on a hydrophilic surface. WT and cda2/cbp1/cda3 conidia, germinated in the presence of 0.01% (w/v) chitosan for 24 hr did not form appressoria. Next, combined treatments of chitosan and IBMX or with HDD were applied. Interestingly, these combinatorial treatments induced appressorium development in the cda2/cbp1/cda3 mutant. IBMX and HDD added in combination did not, however, induce appressorium development in the triple deletion strain. These experiments reveal two important facts: i) Chitosan acts independently of surface hydrophobicity, and ii) Chitosan relays through a separate pathway from IBMX or HDD.
Chitosan is required for germling adhesion
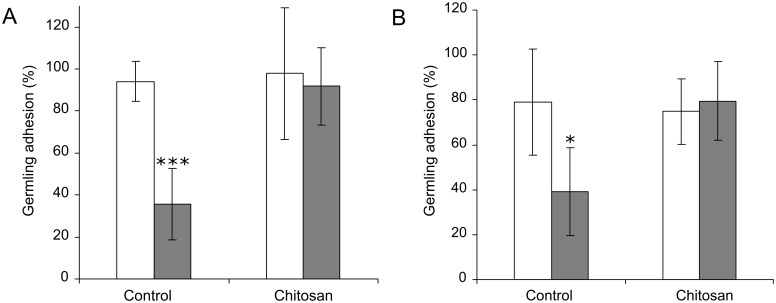
During the course of these assays it was observed that CDA deletion strain germlings washed off artificial surfaces more readily than the WT strain. The percentage of WT and cda2/cbp1/cda3 germlings adhering to hydrophilic and hydrophobic glass surfaces was quantified at 2 hpi, at which point appressorium development had not yet occurred. Germlings of the triple deletion strain demonstrated significantly lower adhesion than the WT, on both surfaces (Student's T-test, p < 0.05) (Fig 12A & 12B). This loss of adhesion could not be explained by lower conidial germination in the cda2/cbp1/cda3 strain; 95% (±2 (SD), n = 3) of WT conidia had germinated by 2 hpi, compared with 98% (±1 (SD), n = 3) in cda2/cbp1/cda3. Remarkably, however, adhesion was fully restored on both surfaces by germinating the conidia in the presence of 0.01% chitosan (w/v), (Fig 12A & 12B). The adhesion of fungal cells to surfaces such as glass or plastic is mediated by cell wall glycoproteins [51]. These can be detected by staining with the fluorescently-labelled mannose-binding lectin Concanavalin A (FITC-ConA). Staining of WT or cda2/cbp1/cda3 germlings at 2 hpi with FITC-ConA revealed that staining intensity was often reduced in cda2/cbp1/cda3 strain (S11 Fig). Quantification of fluorescence intensity revealed significant (p < 0.01) differences between the WT and cda2/cbp1/cda3 germlings in 3 of the 4 experiments (2-way ANOVA with post-hoc Tukey test) (S11 Fig).
Fig 12. Germling adhesion is reduced in the cda2/cbp1/cda3 strain.
Adhesion of WT (white bars) and cda2/cbp1/cda3 germlings (grey bars) on A) hydrophobic and B) hydrophilic glass surfaces with or without 0.01% chitosan (w/v). Conidial suspensions of equal concentrations were inoculated onto each surface and incubated for 2 hr, followed by 5 min washing with H2O, on an orbital shaker at 100 rpm. Germling adhesion was significantly reduced in the triple deletion strain (Student's T-test, *** p < 0.001, * p < 0.05), but restored by the addition of exogenous chitosan. Error bars: SD, n = 4.
Discussion
Chitin deacetylases are a conserved family of enzymes in fungi. They catalyze the removal of acetyl groups from chitin, forming chitosan. It has been hypothesized that the deacetylation of chitin may either act as a 'stealth mechanism' to prevent the activation of a PAMP-triggered immune response in the host plant, or result in crucial alterations to the physical and chemical properties of the cell wall necessary for cellular development and morphogenesis. In this study, we present evidence to refute these hypotheses. Instead, we show that chitin deacetylation during germling morphogenesis is not an absolute requirement for either appressorium development or for pathogenicity. Instead, chitin deacetylation plays a role in perception of physical stimuli during the early stages of germling morphogenesis.
Appressorium development is induced by the perception of physical and chemical stimuli present on the plant surface, with signals then relayed via two key regulatory pathways in M. oryzae. The Pmk1 pathway operates at the earliest stage of appressorium development, and is required for sensing of wax, and possibly for surface attachment. The cAMP pathway, is hypothesized to operate at the commitment phase of appressorial development, and is required for sensing hydrophobicity and cutin monomers [49]. Considerable cross-talk exists between these two pathways, but activation of both is required for appressorium development. Many previous studies have been devoted to characterizing the proteins operating in these pathways (reviewed in [49]). Significantly, deletion of the genes encoding such proteins results in phenotypes that are similar to those reported for the cda mutants. The data from our study, set in the context of previous data on signal perception in M. oryzae [49] allows the role played by chitosan to be elucidated.
When germinated on an artificial hydrophobic surface, conidia of the CDA deletion strains cbp1, cda2/cbp1 and cda2/cbp1/cda3 failed to undergo early differentiation events, and appressorium development was either severely delayed (in cbp1) or rarely occurred at all (in cda2/cbp1 and cda2/cbp1/cda3). Failure to differentiate appressoria on artificial surfaces is a phenotype shared by a number of other deletion strains with putative roles in surface sensing. For example, deletion strains of the signalling mucin Msb2, produce highly elongated, undifferentiated germ tubes on an artificial hydrophobic surface [48], but appressorium development is fully restored when inoculated onto a plant surface, just as in the cda strains described herein. In addition, cutin monomers were also unable to rescue msb2, although the effect of cAMP was not determined. Is there a functional link between Msb2 and chitosan? Msb2 has a single transmembrane domain, which anchors the protein to the plasma membrane, whilst the extracellular portion of the protein resides within the cell wall. This extracellular domain alone can partially suppress the defects associated with MSB2 deletion [52]. Chitosan may be required for cell wall-localization of Msb2; the absence of chitosan may result in loss of Msb2 from the cell wall, resulting in the observed phenotype. However, the precise mechanism by which Msb2 mediates surface sensing remains elusive and this limits speculation on the putative functional link between this protein and chitosan. However, in a recent study, combined deletion of CBP1 and MSB2 had additive effects—virulence and Pmk1 phosphorylation (in vegetative hyphae) were shown to be more reduced in msb2/cbp1 strains than in msb2 strains [52]. Loss of chitosan may therefore affect multiple surface-sensing related processes at the earliest stage of germling morphogenesis, which converge on the Pmk1 pathway. In support of this hypothesis, cbp1 germlings have also been shown to be unable to form hyphopodia on artificial hydrophilic surfaces, a phenotype shared by the pmk1 mutant, but not the cpka mutant [34].
Other proteins with putative roles in surface sensing are the hydrophobin Mpg1 [43, 53], and the putative G-protein coupled receptor Pth11 [54]. The mpg1 and pth11 mutants demonstrate similar defects in appressorium development. That is, germlings produce elongate germ tubes on hydrophobic surfaces, and fail to develop appressoria, as in the cda mutants. There are, however, several important differences in the phenotypes: Firstly, appressorium development is also defective on plant surfaces in mpg1 and pth11. Secondly, germ tubes of mpg1 and pth11 do undergo early differentiation events (germ tube hooking). Thirdly, cAMP can restore appressorium development in both mpg1 and pth11 [53, 54], but not in the cda mutants, although this is complicated by the presence of redundant CDAs that appear to be upregulated by cAMP [55, 56]. This suggests that Mpg1 and Pth11 act upstream of the cAMP pathway, whereas chitosan may not. This hypothesis is supported by the fact that chitosan and IBMX/cutin had synergistic effects on appressorium development on a hydrophilic surface. Yet, the fact that germination in the presence of 1-octacosanol, which acts upstream of the Pmk1 pathway (perceived by the ShoI receptor protein) [48], could only partially restore appressorium development in the cda2/cbp1/cda3 strain, may suggest that activation of the cAMP pathway is also defective in this mutant. On the other hand, cutin monomers, which act through the cAMP pathway [54], did not have a synergistic effect with wax, which may suggest the cAMP pathway is already active in the cda2/cbp1/cda3 strain. Partial rescue with 1-octacosanol could instead be explained by the composition of the wax, which may not effectively mimic the mixture present on leaf surfaces [57].
An alternative explanation for the observed phenotypes in the cda mutants, although one that is not mutually exclusive with those proposed above, is that the lack of germling adhesion is responsible. Germlings of cda2/cbp1/cda3 were much more easily detached than those of the WT, even under relatively gentle washing conditions. This suggests that the adhesive properties of the germ tube are defective in the absence of chitosan. In this scenario, germ-tube localized chitosan is required for the adhesion of the germ tube to the surface itself, and is therefore required for surface sensing, since such sensing mechanisms presumably require close contact between the germ tube and the surface in question. Thus, chitosan may act directly as a polysaccharide adhesin, analagous to the role of galactosaminogalactan (GAG) in the cell wall of Aspergillus fumigatus, which mediates adhesion to hydrophobic surfaces and host cells [58, 59]. Alternatively, chitosan may be required for the attachment of various adhesin-like proteins to the cell wall (or a combination of both). Lack of chitosan in cda2/cbp1/cda3 did result in a reduction of Concanavalin A staining, suggesting a reduction of mannan in the cell wall, but there are several possible explanations for this: i) Chitosan is directly required for attachment of mannoproteins to the cell wall, ii) Loss of chitosan results in large-scale changes in cell wall composition, affecting attachment of mannoproteins or resulting in a reduction in non-protein associated mannan, iii) Mannoproteins are produced, but secretion to the outer wall is affected by loss of chitosan or iv) Lack of surface sensing and activation of Pmk1 and/or cAMP pathways means that production of the mannoproteins does not occur in the first place, i.e. reduced germling adhesion is a consequence, and not the cause of defective surface sensing. Further investigations are required to distinguish between these possibilities.
The rescue of appressorium development in the cda mutants by exogenous chitosan and its association with the cell wall was an unexpected result, and it is unclear exactly how this occurs. Because the charge of the exogenous chitosan was of great importance, this may indicate that an electrostatic interaction is responsible for its association with the cell wall. This, in turn, suggests the existence of an anionic cell wall component, which may form a complex with the cationic chitosan. At present, little is known of anionic cell wall components in fungi. Uronic acids are known to be present in the cell walls of some fungi [60–63], as is phosphomannan [64–66], but their presence in M.oryzae has not been investigated. In addition to an electrostatic association, the possibility of enzymatic incorporation of exogenous chitosan must also be considered. CRH transglycosylases have previously been shown to be able to incorporate exogenous fluorescently-labelled oligosaccharides in S.cerevisiae [67]. It is therefore possible that other enzymes with similar activities may be responsible for the hypothesized incorporation of exogenous chitosan.
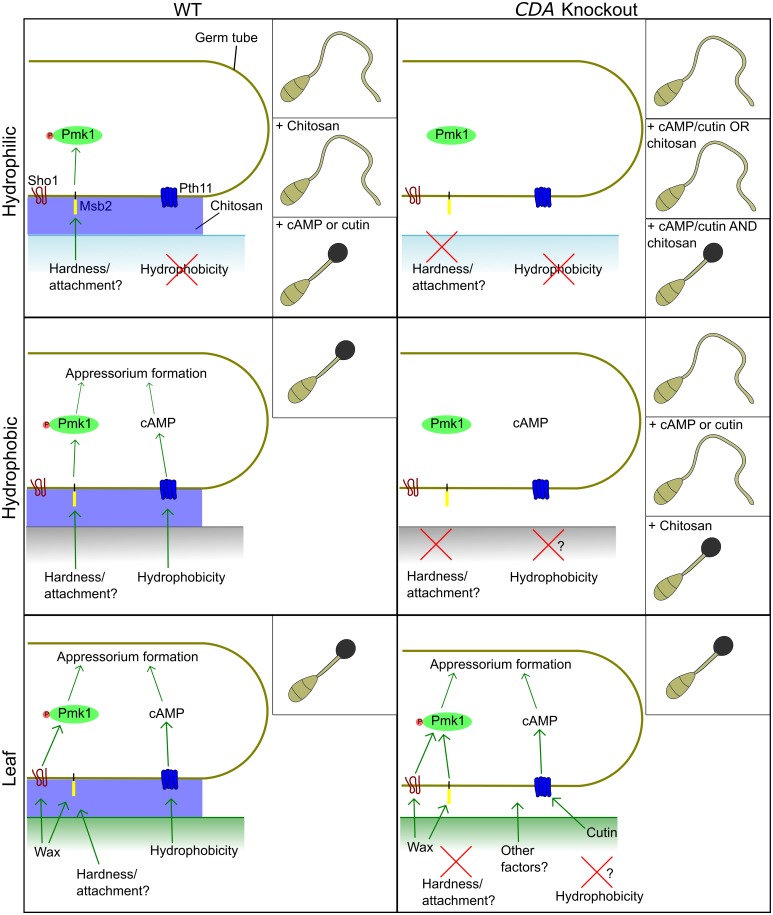
The role of chitosan in appressorium development is an intriguing one, and there remain several unresolved issues, as discussed above. Despite this, the model presented in Fig 13 is consistent with all of the data presented herein, and with previous studies characterizing surface sensing proteins in M.oryzae. In this model, germ-tube localized chitosan is required for the activation of the Pmk1 MAP kinase pathway in response to a surface, but independently of surface hydrophobicity. Optimal activation of the cAMP pathway may also require chitosan, but it is not possible to conclusively determine whether just one or both of the signalling pathways are defective in cda mutants from the current data.
Fig 13. Model describing the role of chitosan in appressorium development.
Germling development on three different surfaces is shown (hydrophilic, hydrophobic and leaf), with observed phenotypes shown inset. Successful appressorium development is dependant upon activation of both the Pmk1 and cAMP pathways. The absence of chitosan prevents complete activation of the Pmk1 pathway, and possibly the cAMP pathway. The addition of chitosan and/or chemical inducers of these pathways restores appressorium development in the CDA deletion strains. Thus, germ tube localized chitosan likely acts upstream of both the Pmk1 and cAMP pathways to induce appressorium development.
The deacetylation of chitin was hypothesized to cause profound changes to the chemical and physical properties of the cell wall, which could be crucial for cellular morphogenesis. However, no evidence for this has been found in the present study. Germlings of the cda2/cbp1/cda3 strain developed appressoria that were morphologically indistinguishable from the WT strain on plant surfaces, despite the absence of detectable chitosan. Additionally, appressorium development in cda2/cbp1/cda3 was restored by a short incubation with FITC-chitosan that was only incorporated into the germ tube. Not only was chitosan not required for morphogenesis of appressoria, but appressorium function also seemed to be unaffected by the absence of chitosan, since plant penetration continued to be achieved successfully. Chitosan is therefore also unlikely to be a key structural component of the cell wall, in contrast to Cryptococcus neoformans where loss of chitosan impaired cellular integrity [31]. On the other hand, it is also possible that compensatory changes in cell wall composition occurred in the absence of chitosan, which were not investigated. The upregulation of other cell wall components may be sufficient to maintain cell wall integrity in the cda mutants. In a similar vein, it is not inconceivable that compensatory changes were also responsible for allowing appressorium morphogenesis to occur in the absence of chitosan i.e. there are multiple, redundant morphogenic mechanisms operating during appressorium development.
A second hypothesized role of chitosan is in the protection from plant chitinases. However, the findings presented here, together with evidence from previous studies do not support this role for chitosan, at least in germlings. Germlings of cda2/cbp1/cda3 in which chitosan was absent remained intact on leaf surfaces, in contrast to those of the ags mutant which lack α1,3-glucan and were destroyed, presumably by degradative enzymes on the leaf surface [18]. This suggests that α1,3-glucan may be the cell wall component with the protective role in M.oryzae germlings. It would also be valuable to investigate the hypothesized protective effects of chitin deacetylation during in planta growth. Chitosan staining was not performed on invasive hyphae in the cda mutants, although qRT-PCR analysis suggested that the chitin deacetylases operating during appressorium development are not highly expressed during invasive growth (S2 Fig). Instead, CDA1 and CDA6 appear to be involved in chitin deacetylation at this stage of infection. Further investigations should therefore focus on the characterization of these genes to determine the role of chitosan in invasive hyphae.
The deacetylation of chitin is hypothesized to require a degree of collaboration between the membrane-localized chitin synthases, and cell wall or membrane-associated chitin deacetylases [31, 68]. Evidence from this study may lend further support to this hypothesis. Deletion of CHS7 results in an almost identical phenotype to multiple CDA deletion [69], suggesting that Cbp1, Cda2 and Cda3 may deacetylate the chitin synthesized by Chs7. Further investigation is required to determine the relationship between chitin synthesis and deacetylation during appressorium development in M.oryzae.
In summary, the investigation of chitin deacetylation during appressorium development in M.oryzae has yielded unexpected and intriguing data. The deacetylation of chitin by at least three chitin deacetylases, with overlapping roles, is required for surface sensing in germlings. Yet, this requirement is surface dependant, due to the multiple, redundant mechanisms by which appressorium development can be induced in M.oryzae. Nevertheless, this study provides a novel insight into the mechanisms behind the perception of physical stimuli in M.oryzae, and also demonstrates a novel way in which the cell wall is crucial in acting as an interface between fungal cells and their environment. Importantly, evidence in support of the long-standing hypothesis regarding the role of chitin deacetylation was not found; chitosan is not required as a 'stealth' mechanism in germlings of M.oryzae, and so the role of this cell wall component in the interactions between phytopathogenic fungi and their hosts may need to be reconsidered. However, further work is required to determine whether or not chitosan acts as a 'stealth mechanism' during in planta growth of M.oryzae, as this was not determined in this study.
Materials and Methods
Fungal strains and growth conditions
The wild-type (WT) rice pathogenic Magnaporthe oryzae strain Guy11 and mutant strains were cultured at 24°C with a 14 h light 10 h dark cycle. Strain maintenance and composition of media were essentially as described by Talbot et al [43].
RT-PCR
RNA was extracted from rice leaves inoculated with conidia of the Guy11 strain using the Qiagen RNeasy RNA extraction kit, according to manufacturer's instructions. RNA concentration was determine using a ThermoScientific ND-1000 NanoDrop spectrophotometer, integrity was evaluated by gel electrophoresis. Genomic DNA was removed by using an RNase free DNase set (Qiagen), according to manufacturers’ instructions. Reverse transcription of 1 μg of RNA into cDNA was performed by using the Maxima First Strand cDNA synthesis kit (ThermoFisher Scientific), according to manufacturers instructions, using random hexamer primers. qRT-PCR was performed with Power SYBR Green PCR master mix (ThermoFisher Scientific), on an Applied Biosystems 7300 Real-time PCR system. Primers are listed in S2 Table (37–58). Primers were designed to span an intron where possible (not all CDA genes have introns), and efficiency of the primers was 85–104% (average efficiency 93.6%). No amplification was observed in samples that were not reverse transcribed (-RT control), in samples without RNA (NTC control), or in RNA extracted from mock-inoculated leaves. Relative transcript abundance was calculated by the efficiency correction method [70] as follows: Abundance = Etarget (Ct(reference)-Ct(target)).
Targeted deletion of M. oryzae CDAs
Single CDA deletion strains were generated by replacing the coding sequences of CBP1 (MGG_12939), CDA2 (MGG_09159) and CDA3 (MGG_04172) with a hygromycin resistance cassette [71]. Briefly, sequences flanking the target genes (~1.5kb upstream, and ~1.1kb downstream) were amplified by PCR, using primers 7–10 (for CBP1), 13–16 (for CDA2) and 17–20 (For CDA3) (primers are listed in S2 Table). These fragments were joined to the hygromycin resistance gene by overlapping PCR, using primer pairs 7/4 and 3/10 (for the CBP1 deletion construct), 13/4 and 3/16 (for the CDA2 deletion construct), 17/4 and 3/20 (for the CDA3 deletion construct). The final, complete construct was made by overlapping PCR, amplifying the products from the previous reactions using primer pairs 7/10 (for CBP1), 13/16 (for CDA2) and 17/20 (for CDA3). The final PCR product was used directly for DNA-mediated protoplast transformation of WT Guy11 strain following protocols described by Talbot et al (39). Putative transformants were selected on minimal medium (MM) supplemented with 300 μg ml-1 hygromycin B (Calbiochem, Merck, Darmstadt, Germany). Deletion of the target gene was confirmed by both PCR and Southern Blot analysis, as described in Samalova et al [72].
Double CDA deletion strains were generated in the cda2 or cbp1 background strains. The coding sequences of CBP1 and CDA3 were replaced with a bialaphos resistance cassette. The deletion construct was made as described above, except primers 8 & 9 were substituted for 11 & 12 (for CBP1), and primers 18 & 19 for 21 & 22 (for CDA3). The CBP1 deletion cassette was transformed directly into protoplasts of the cda2 strain, to generate the cda2/cbp1 mutant. The CDA3 deletion cassette was transformed into both the cda2 and cbp1 background strains, to generate the cda2/cda3 and cbp1/cda3 mutants. Putative transformants were selected on defined complex medium (DCM) supplemented with 60 μg ml-1 Bialophos (Goldbio, St Louis, MO, USA). Deletion strains were confirmed as above.
To generate the triple cda2/cbp1/cda3 mutant, CDA3 was deleted in the cda2/cbp1 mutant. In this case, the coding sequence of CDA3 was replaced by a sulphonylurea resistant allele of the M.oryzae ILV gene (MGG_06868). Since the sulphonylurea transformation constructs were too large to be generated by over-lapping PCR, GAP-repair S. cerevisiae cloning [73] was used to assemble the constructs in pNEB1284 vector. Primers 5,6 and 23–26 were used to amplify the required DNA fragments. Putative transformants were selected on BDCM medium supplemented with 100 μg ml-1 chlorimuron ethyl (Sigma Aldrich, UK) and confirmed as specified above.
Cloning of fluorescently tagged CDAs
Standard molecular techniques [74] were used to prepare the complementation constructs with fluorescently tagged CDAs. A set of transformation vectors based on pUCAP was generated as described in Samalova et al. [75]. The vectors contain polyadenylation signal pATrpC and either bialophos or hygromycin resistance marker that was cloned into re-created SalI sites using primer pairs 1/2 or 3/4 respectively (see S2 Table and S12 Fig). For PCR amplification of CBP1 and CDA2, primer pairs 27/28 and 29/30 were used, respectively. Genomic DNA from the WT strain Guy11 was used as a template, and amplified using Herculase DNA polymerase (Agilent). This resulted in amplification of the coding sequence of the genes (without stop codons), together with 2 kb of native promoter sequence for CBP1 and 1.3 kb for CDA2. The PCR products were cloned into the AscI sites of the vector described above (S12 Fig), creating C-terminal mCherry fusions.
Confocal imaging
Conidia (2.5 x 105 ml -1) of Guy11 and complemented strains were collected from 10 day old plates and inoculated in 50 μl droplets onto hydrophobic glass cover-slip,; onion peels, or rice leaf sheaths; as described in Samalova et. al., [75] and incubated for specified times in the growth chamber. For viewing mCherry fluorescence, the samples were viewed using the C-Apochromat 40x/1.2 water corrected objective lens of a Zeiss LSM 510 Meta confocal microscope at 543 nm excitation from the HeNe laser and emission collected with BP565-615 filter for mCherry.
Calcofluor White staining was performed as follows: Conidia (2.5 x 105 ml -1) of the Guy11 and mutant strains were inoculated in 50 μl droplets onto hydrophobic glass cover-slips, and incubated for specified times in the growth chamber. After the incubation, the water droplet was removed from the cover-slip, and replaced with 100 μl of 0.05% Calcofluor White solution (Sigma-Aldrich, UK), and incubated for 20 min. Samples were then washed briefly with dH2O, and viewed using the Zeiss LSM510 microscope as above, with 405 nm excitation and emission collected with an LP420 filter.
OGA488 staining was performed essentially as described in [36]. Briefly, Conidia (2.5 x 105 ml-1) of the Guy11 and mutant strains were inoculated in 50 μl droplets onto hydrophobic glass cover-slips, onion epidermis, or rice leaf sheaths and incubated for specified times in the growth chamber. Samples were washed briefly with 25 mM MES (pH 5.6), and incubated with OGA488 (a generous gift from William Willats [36]) (diluted 1/1000 in 25 mM MES) for 15 min on ice. This was followed by 2–3 brief washes with 25 mM MES, after which the samples were viewed using the C-Apochromat 40x/1.2 water corrected objective lens of a Zeiss LSM 510 Meta confocal microscope, at 488 nm excitation and emission collected with an LP505 filter.
Staining of germlings with FITC-labelled Concanavalin A (ConA-FITC) (Sigma-Aldrich, UK) was performed as follows: Conidia (2.5 x 105 ml-1) of the Guy11 and mutant strains were inoculated in 50 μl droplets onto hydrophobic glass cover-slips, and incubated in the growth chamber for 2 hr. Cover-slips were washed briefly with PBS, then incubated with 40 μg/ml ConA-FITC for 20 min on ice, then washed briefly with PBS 2–3 times. Samples were viewed using the C-Apochromat 40x/1.2 water corrected objective lens of a Zeiss LSM 510 Meta confocal microscope, at 488 nm excitation and emission collected with an LP505 filter. Fluorescence intensity was quantified using ImageJ.
Staining with the monoclonal anti-chitosan antibody mAbG7 (a generous gift from Stefan Schillberg [35]) was performed as follows. Conidia (2.5 x 105 ml-1) of the Guy11 strain were inoculated in 50 μl droplets onto hydrophobic glass cover-slips, and incubated in the growth chamber for the specified time. Samples were first blocked by incubation with 2% BSA (w/v) in PBS for 1 hr at room temperature. Samples were washed 3 times with PBS/T (PBS +0.05% Tween 20), for 5 min each on an orbital shaker (~70 rpm) and then incubated with the primary antibody (mAbG7, at 10 μg/ml in PBS) for 1.5 hr at room temperature. This incubation was followed by 3 more washing steps as described above, and incubation with the secondary antibody (FITC-labelled anti Mouse IgM, 5 μg/ml in PBS) at room temperature for a further 1.5 hr. Finally, the secondary antibody was removed by 3 more washing steps with PBS/T as described previously and viewed under using the Zeiss LSM510 microscope, as described above for ConA-FITC staining. A negative control was included in all experiments, in which samples were only incubated with the secondary antibody.
Staining with the polyclonal anti-chitosan antibody (a generous gift from Holger Deising [19]) was performed as for the mAbG7, except that the antibody was used at a dilution of 1/100 from the original antiserum, and the secondary antibody (a FITC-labelled anti-rabbit IgG) was used at 10 μg/ml (in PBS).
Pathogenicity and infection-related morphogenesis assays
Conidial germination and appressorium development were assessed at 1, 8, 16 or 24 hpi by following germling differentiation on hydrophobic glass cover-slips (Gerhard Menzel, Glasbearbeitungswerk GmbH & Co., Braunschweig, Germany). Conidia (2.5 x 105 ml-1) of the Guy11 and mutant strains were inoculated in 50 μl droplets onto hydrophobic glass cover-slips, and incubated in the growth chamber for the specified time. Samples were viewed under an Olympus BX50 microscope, and ~500 germlings in 3 independent experiments counted per strain/timepoint. For germinations in the presence of chemical inducers of appressorium development, IBMX (Sigma-Aldrich, UK) was used at 2.5 mM (from a 250 mM stock in DMSO), 1,16 hexadecanediol (Sigma-Aldrich, UK) at 200 μM (from a 50 mM stock in ethanol) and diacylglycerol (1,2-dioctanoyl-sn-glycerol) (Enzo Life Sciences) at 58 μM (from a 7.25 mM stock in DMSO). Chitosan and its derivatives were used at a final concentration of 0.01% or 0.001%, from a 1% stock (except for FITC-chitosan which was from a 0.08% stock). For germinations in the presence of wax (1-octacosanol) (Sigma-Aldrich, UK), the wax was first dissolved in chloroform to a concentration of 4 mg/ml. In a laboratory fume hood, 100 μl of this stock was pipetted onto a hydrophobic glass coverslip and the chloroform allowed to evaporate, leaving a layer of wax on the coverslip. Conidia were then inoculated onto this surface, as described above. For germinations on hydrophilic glass coverslips (Heathrow Scientific) the protocol was identical except that only 20 μl of conidial suspension was used. Coverslips were placed in square Petri dishes with damp filter paper and sealed with Parafilm to prevent evaporation.
Cuticle penetration was assessed by scoring the frequency with which appressoria formed penetration pegs and intracellular infection hyphae on rice leaf sheaths, after incubation at 24°C for 24 h.
Leaf infection assays were performed on blast-susceptible, 21-day-old seedlings of rice (Oryza sativa L.) cultivar CO39. Assays on detached leaves were performed as described in [72]. For assays on whole plants, 21-day-old seedlings of rice (Oryza sativa L.) cultivar CO39 were spray inoculated with 4 ml of conidial suspension at three different concentrations (1.25 x 105, 6.25 x 104 or 3.13 x 104 conidia/ml, in 0.2% (w/v) gelatine water). A mock inoculation of 0.2% (w/v) gelatine water was included as a negative control. Infection was assessed 4 days later.
Germling adhesion assay
Adhesion assays were performed on both hydrophobic and hydrophilic glass coverslips. 20 μl droplets of conidial suspension (either with or without 0.01% chitosan) of each strain were pipetted onto the coverslips, which were placed into a humidity chamber and incubated at 24°C for 2 hr to allow germination. At 2 hpi, half of the coverslips were removed and placed into a 50 mm Petri dish containing 5 ml dH2O, and shaken on an orbital shaker at 100 rpm for 5 min. Coverslips were then removed and mounted on slides for viewing under the 10X objective lens of an Olympus BX50 microscope. Conidia were counted from one field of view (for hydrophobic coverslips) or three fields of view (for hydrophilic coverslips), with a conscious effort made to locate the area of the coverslip with the highest conidial density. For each strain, the number of conidia counted on the washed coverslips was compared with those counted on the unwashed coverslips, and percentage conidial adhesion calculated as: 100—((conidia on unwashed coverslip—conidia on washed coverslip) /conidia on unwashed coverslip) x 100.
Synthesis of FITC-chitosan
FITC-labelled chitosan was synthesized exactly as described previously [45].
Supporting Information
Conserved active site residues are marked in green boxes, and the zinc binding triad in blue boxes.
(TIF)
A) Development of appressoria (black arrow head) at 5 hpi on rice leaves. B) Melanized appressoria (white arrow head) and invasive hyphae (black arrow head) at 36 hpi on rice leaves. Scale bars: 20 μm. C) Expression of CDAs during in planta development at 36 hpi. Error bars show standard deviation, n = 3.
(TIF)
A) Schematic of deletion strategy, whereby the gene of interest is replaced by a gene encoding antibiotic resistance. Arrows show position of primers used to screen transformants. Pair 1 amplifies the gene of interest, confirming its absence in the deletion strain. Pairs 2 and 3 test for integration of the deletion construct at the desired locus. B) PCR analysis of single CDA deletion strains, showing successful deletion of CBP1, CDA2 and CDA3. C) PCR analysis of double CDA deletion strains (gene tested for written in red). D) PCR analysis of triple CDA deletion strain, showing successful deletion of CDA3. Note: Some of the gel pictures are composites images of different parts of the same gel.
(TIF)
A) Targeted gene deletion strategy, whereby the gene of interest is replaced by a gene encoding resistance to an antibiotic, due to homologous recombination between sequences (green). Southern blots showing successful single B), double C) and triple D) knockouts of chitin deacetylases.
Blots containing restriction digested gDNA of putative deletion strains were hybridised with α-32P labelled DNA homologous to the hygromycin, bialaphos or sulphonylurea resistance genes respectively. The cartoon above each blot shows the expected band size based upon the position of the restriction enzyme sites at each locus. Successful gene replacement is evidenced by a single band of the expected size, (as calibrated against standard gel size marker ladder and with band sizes given in kilobases (kb). For the double deletion strains, cross-hybridisation was observed between the hygromycin and bialaphos resistance cassettes, due to a common promoter sequence. These faint bands are labelled in B.
(TIF)
Conidia of the WT and cda2/cbp1/cda3 strains were inoculated onto the surfaces in 50 μl droplets, and incubated for 24 hr. Scale bars: 40 μm.
(TIF)
A) OGA488 staining of WT, cda2/cbp1 and cda2/cbp1/cda3 germlings at 24 hpi, showing absence of labelling in appressoria of the deletion strains (white arrow heads). B) Calcofluor white staining of germlings at 16 hpi, showing abnormally elongated, septate germ tubes (white arrow heads) in mutant strains, and abnormally shaped appressoria in cbp1. Scale bars: 20 μm.
(TIF)
A) Cbp1:mCherry and B) Cda2:mCherry fluorescence at later stages of appressorium development (as indicated). Scale bars: 10 μm.
(TIF)
Molecular structure, degree of polymerization and degree of deacetylation (DDA) are indicated (when known).
(TIF)
A) Pathogenicity of the cda2/cbp1/cda3 deletion strain on rice leaves, showing comparable lesion density to the WT strain (two independent triple deletion lines are shown). A mock inoculation of 0.2% gelatine was included as a negative control. Scale bars: 1 cm. B) Quantification of lesion numbers on detached rice leaves inoculated with different cda mutant strains, normalized to the Guy11 WT strain (± SD, n = 3).
(TIF)
Appressorium development was observed in cda2/cbp1/cda3 (white arrow heads) in the presence of 1-octacosanol, but germ tubes remained abnormally elongated. Scale bars: 50 μm.
(TIF)
A) Germlings of the WT and cda2/cbp1/cda3 strain, stained with FITC-ConA. Pictures were taken from Experiment 1 (quantified in B). Scale bars: 50μm. B) Fluorescence intensity of germlings stained with FITC-ConA. Fluorescence intensities of ~140 germ tubes were measured, across 4 independent experiments. White bars: WT, grey bars: cda2/cbp1/cda3. Significant differences were found in 3 of the 4 experiments (2-way ANOVA with post-hoc Tukey test (** = p <0.01, *** = p < 0.001))
(TIF)
(TIF)
(XLS)
(XLS)
Conidia of the WT and cda mutants were inoculated onto an artificial surface and incubated for 16 hr, in the presence of IBMX (3-isobutyl-1-methylxanthine), HDD (1,16 hexadecanediol; cutin monomer) or DAG (1,2-dioctanoyl-sn-glycerol). Significant (Mann-Whitney U-test, p < 0.001) reductions (asterisks) in germ tube lengths were observed in the deletion strains with all treatments, when compared with the control. ND—Not determined.
(TIF)
Acknowledgments
We are grateful to Dr. Marketa Samalova for her advice and help, and Sarah Rodgers and Caroline O'Brien for their technical assistance. Charlotte Kirchhelle provided invaluable help with fluorescence quantification and statistical analysis. We thank Stefan Schillberg for supplying the mAbG7 monoclonal antibody, Holger Deising for supplying the polyclonal antichitosan antibody, and William Willats for supplying the OGA488.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
IAG was funded by BBSRC CASE studentship; SJG gratefully acknowledges BBSRC grant award BB/J008923/1. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Latge J-P. The cell wall: a carbohydrate armour for the fungal cell. Molecular Microbiology. 2007;66(2):279–90. 10.1111/j.1365-2958.2007.05872.x WOS:000249834700001. [DOI] [PubMed] [Google Scholar]
- 2. Latge J-P, Beauvais A. Functional duality of the cell wall. Current Opinion in Microbiology. 2014;20:111–7. 10.1016/j.mib.2014.05.009 WOS:000340991400019. [DOI] [PubMed] [Google Scholar]
- 3. Cabib E, Arroyo J. How carbohydrates sculpt cells: chemical control of morphogenesis in the yeast cell wall. Nature reviews Microbiology. 2013;11(9):648–55. 10.1038/nrmicro3090 MEDLINE:23949603. [DOI] [PubMed] [Google Scholar]
- 4. Bowman SM, Free SJ. The structure and synthesis of the fungal cell wall. Bioessays. 2006;28(8):799–808. 10.1002/bies.20441 WOS:000240394500005. [DOI] [PubMed] [Google Scholar]
- 5. Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease. Trends in Biotechnology. 2009;27(3):141–50. 10.1016/j.tibtech.2008.12.002 WOS:000264279000004. [DOI] [PubMed] [Google Scholar]
- 6. Perez-Nadales E, Nogueira MFA, Baldin C, Castanheira S, El Ghalid M, Grund E, et al. Fungal model systems and the elucidation of pathogenicity determinants. Fungal Genetics and Biology. 2014;70:42–67. 10.1016/j.fgb.2014.06.011 WOS:000341617000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilbert RD, Johnson AM, Dean RA. Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiological and Molecular Plant Pathology. 1996;48(5):335–46. WOS:A1996UT36200004. [Google Scholar]
- 8. Howard RJ, Ferrari MA, Roach DH, Money NP. Penetration of hard substrates by a fungus employing enormous turgor pressures. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(24):11281–4. 10.1073/pnas.88.24.11281 WOS:A1991GV87700062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fujikawa T, Kuga Y, Yano S, Yoshimi A, Tachiki T, Abe K, et al. Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Molecular Microbiology. 2009;73(4):553–70. 10.1111/j.1365-2958.2009.06786.x WOS:000268793000003. [DOI] [PubMed] [Google Scholar]
- 10. Oliveira-Garcia E, Valent B. How eukaryotic filamentous pathogens evade plant recognition. Current Opinion in Microbiology. 2015;26:92–101. 10.1016/j.mib.2015.06.012 WOS:000362132300016. [DOI] [PubMed] [Google Scholar]
- 11. Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant Journal. 2010;64(2):204–14. 10.1111/j.1365-313X.2010.04324.x WOS:000282876100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(3):E404–E13. 10.1073/pnas.1312099111 WOS:000329928400015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–9. 10.1038/nature05286 WOS:000242018300039. [DOI] [PubMed] [Google Scholar]
- 14. Torres MA, Jones JDG, Dangl JL. Reactive oxygen species signaling in response to pathogens. Plant Physiology. 2006;141(2):373–8. 10.1104/pp.106.079467 WOS:000238168800011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schwessinger B, Zipfel C. News from the frontline: recent insights into PAMP-triggered immunity in plants. Current Opinion in Plant Biology. 2008;11(4):389–95. 10.1016/j.pbi.2008.06.001 WOS:000258852300006. [DOI] [PubMed] [Google Scholar]
- 16. de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, et al. Conserved Fungal LysM Effector Ecp6 Prevents Chitin-Triggered Immunity in Plants. Science. 2010;329(5994):953–5. 10.1126/science.1190859 WOS:000281084800037. [DOI] [PubMed] [Google Scholar]
- 17. Mentlak TA, Kombrink A, Shinya T, Ryder LS, Otomo I, Saitoh H, et al. Effector-Mediated Suppression of Chitin-Triggered Immunity by Magnaporthe oryzae Is Necessary for Rice Blast Disease. Plant Cell. 2012;24(1):322–35. 10.1105/tpc.111.092957 WOS:000300881800027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fujikawa T, Sakaguchi A, Nishizawa Y, Kouzai Y, Minami E, Yano S, et al. Surface alpha-1,3-Glucan Facilitates Fungal Stealth Infection by Interfering with Innate Immunity in Plants. Plos Pathogens. 2012;8(8). 10.1371/journal.ppat.1002882 WOS:000308558000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. El Gueddari NE, Rauchhaus U, Moerschbacher BM, Deising HB. Developmentally regulated conversion of surface-exposed chitin to chitosan in cell walls of plant pathogenic fungi. New Phytologist. 2002;156(1):103–12. 10.1046/j.1469-8137.2002.00487.x WOS:000177962000012. [DOI] [Google Scholar]
- 20. Pochanavanich P, Suntornsuk W. Fungal chitosan production and its characterization. Letters in Applied Microbiology. 2002;35(1):17–21. 10.1046/j.1472-765X.2002.01118.x WOS:000176439300003. [DOI] [PubMed] [Google Scholar]
- 21. Iriti M, Faoro F. Chitosan as a MAMP, searching for a PRR. Plant signaling & behavior. 2009;4(1):66–8. MEDLINE:19704712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brunner F, Stintzi A, Fritig B, Legrand M. Substrate specificities of tobacco chitinases. Plant Journal. 1998;14(2):225–34. WOS:000073794300010. [DOI] [PubMed] [Google Scholar]
- 23. Ruiz-Herrera J, Ortiz-Castellanos L. Analysis of the phylogenetic relationships and evolution of the cell walls from yeasts and fungi. Fems Yeast Research. 2010;10(3):225–43. 10.1111/j.1567-1364.2009.00589.x WOS:000276604700001. [DOI] [PubMed] [Google Scholar]
- 24. Lenardon MD, Munro CA, Gow NAR. Chitin synthesis and fungal pathogenesis. Current Opinion in Microbiology. 2010;13(4):416–23. 10.1016/j.mib.2010.05.002 WOS:000281494500005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christodoulidou A, Bouriotis V, Thireos G. Two sporulation-specific chitin deacetylase-encoding genes are required for the ascospore wall rigidity of Saccharomyces cerevisiae. Journal of Biological Chemistry. 1996;271(49):31420–5. WOS:A1996VW68600063. [DOI] [PubMed] [Google Scholar]
- 26. Mishra C, Semino CE, McCreath KJ, DelaVega H, Jones BJ, Specht CA, et al. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast. 1997;13(4):327–36. WOS:A1997WW94600004. [DOI] [PubMed] [Google Scholar]
- 27. Christodoulidou A, Briza P, Ellinger A, Bouriotis V. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. Febs Letters. 1999;460(2):275–9. 10.1016/s0014-5793(99)01334-4 WOS:000083504800017. [DOI] [PubMed] [Google Scholar]
- 28. Suda Y, Rodriguez RK, Coluccio AE, Neiman AM. A Screen for Spore Wall Permeability Mutants Identifies a Secreted Protease Required for Proper Spore Wall Assembly. Plos One. 2009;4(9). 10.1371/journal.pone.0007184 WOS:000270176200017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lickfeld M, Schmitz H-P. A network involving Rho-type GTPases, a paxillin and a formin homologue regulates spore length and spore wall integrity in the filamentous fungus Ashbya gossypii. Molecular Microbiology. 2012;85(3):574–93. 10.1111/j.1365-2958.2012.08128.x WOS:000306685300012. [DOI] [PubMed] [Google Scholar]
- 30. Banks IR, Specht CA, Donlin MJ, Gerik KJ, Levitz SM, Lodge JK. A chitin synthase and its regulator protein are critical for chitosan production and growth of the fungal pathogen Cryptococcus neoformans. Eukaryotic Cell. 2005;4(11):1902–12. 10.1128/ec.4.11.1902-1912.2005 WOS:000233368100015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Baker LG, Specht CA, Donlin MJ, Lodge JK. Chitosan, the deacetylated form of chitin, is necessary for cell wall integrity in Cryptococcus neoformans. Eukaryotic Cell. 2007;6(5):855–67. 10.1128/ec.00399-06 WOS:000246718300010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baker LG, Specht CA, Lodge JK. Cell Wall Chitosan Is Necessary for Virulence in the Opportunistic Pathogen Cryptococcus neoformans. Eukaryotic Cell. 2011;10(9):1264–8. 10.1128/ec.05138-11 WOS:000294453000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kamakura T, Yamaguchi S, Saitoh K, Teraoka T, Yamaguchi I. A novel gene, CBP1, encoding a putative extracellular chitin-binding protein, may play an important role in the hydrophobic surface sensing of Magnaporthe grisea during appressorium differentiation. Molecular Plant-Microbe Interactions. 2002;15(5):437–44. 10.1094/mpmi.2002.15.5.437 WOS:000175529100004. [DOI] [PubMed] [Google Scholar]
- 34. Tucker SL, Besi MI, Galhano R, Franceschetti M, Goetz S, Lenhert S, et al. Common Genetic Pathways Regulate Organ-Specific Infection-Related Development in the Rice Blast Fungus. Plant Cell. 2010;22(3):953–72. 10.1105/tpc.109.066340 WOS:000276983900031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schubert M, Agdour S, Fischer R, Olbrich Y, Schinkel H, Schillberg S. A Monoclonal Antibody That Specifically Binds Chitosan In Vitro and In Situ on Fungal Cell Walls. Journal of Microbiology and Biotechnology. 2010;20(8):1179–84. WOS:000281405200002. [DOI] [PubMed] [Google Scholar]
- 36. Mravec J, Kračun SK, Rydahl MG, Westereng B, Miart F, Clausen MH, et al. Tracking developmentally regulated post-synthetic processing of homogalacturonan and chitin using reciprocal oligosaccharide probes. Development. 2014;141(24):4841–50. 10.1242/dev.113365 . [DOI] [PubMed] [Google Scholar]
- 37. Lee YH, Dean RA. Hydrophobicity of contact surface induces appressorium formation in Magnaporthe grisea . Fems Microbiology Letters. 1994;115(1):71–5. 10.1111/j.1574-6968.1994.tb06616.x WOS:A1994MY20200012. [DOI] [Google Scholar]
- 38. Blair DE, Hekmat O, Schuttelkopf AW, Shrestha B, Tokuyasu K, Withers SG, et al. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry. 2006;45(31):9416–26. 10.1021/bi0606694 WOS:000239422400006. [DOI] [PubMed] [Google Scholar]
- 39. Hekmat O, Tokuyasu K, Withers SG. Subsite structure of the endo-type chitin deacetylase from a Deuteromycete, Colletotrichum lindemuthianum: an investigation using steady-state kinetic analysis and MS. Biochemical Journal. 2003;374:369–80. 10.1042/bj20030204 WOS:000185317800010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systems Biology. 2011;7 10.1038/msb.2011.75 WOS:000296652600007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Soanes DM, Chakrabarti A, Paszkiewicz KH, Dawe AL, Talbot NJ. Genome-wide Transcriptional Profiling of Appressorium Development by the Rice Blast Fungus Magnaporthe oryzae. Plos Pathogens. 2012;8(2). 10.1371/journal.ppat.1002514 WOS:000300728100022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dong Y, Li Y, Zhao M, Jing M, Liu X, Liu M, et al. Global Genome and Transcriptome Analyses of Magnaporthe oryzae Epidemic Isolate 98–06 Uncover Novel Effectors and Pathogenicity-Related Genes, Revealing Gene Gain and Lose Dynamics in Genome Evolution. Plos Pathogens. 2015;11(4). 10.1371/journal.ppat.1004801 WOS:000354978000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea . Plant Cell. 1993;5(11):1575–90. 10.1105/tpc.5.11.1575 WOS:A1993MK55500010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beckerman JL, Ebbole DJ. MPG1, a gene encoding a fungal hydrophobin of Magnaporthe grisea, is involved in surface recognition. Molecular Plant-Microbe Interactions. 1996;9(6):450–6. 10.1094/mpmi-9-0450 WOS:A1996UZ73300003. [DOI] [PubMed] [Google Scholar]
- 45. Qaqish RB, Amiji MM. Synthesis of a fluorescent chitosan derivative and its application for the study of chitosan-mucin interactions. Carbohydrate Polymers. 1999;38(2):99–107. 10.1016/s0144-8617(98)00109-x WOS:000078497600001. [DOI] [Google Scholar]
- 46. Saitoh H, Fujisawa S, Mitsuoka C, Ito A, Hirabuchi A, Ikeda K, et al. Large-Scale Gene Disruption in Magnaporthe oryzae Identifies MC69, a Secreted Protein Required for Infection by Monocot and Dicot Fungal Pathogens. Plos Pathogens. 2012;8(5). 10.1371/journal.ppat.1002711 WOS:000305322900044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qin B-X, Tang D, Huang J, Li M, Wu X-R, Lu L-L, et al. Rice OsGL1-1 Is Involved in Leaf Cuticular Wax and Cuticle Membrane. Molecular Plant. 2011;4(6):985–95. 10.1093/mp/ssr028 WOS:000297377100005. [DOI] [PubMed] [Google Scholar]
- 48. Liu W, Zhou X, Li G, Li L, Kong L, Wang C, et al. Multiple Plant Surface Signals are Sensed by Different Mechanisms in the Rice Blast Fungus for Appressorium Formation. Plos Pathogens. 2011;7(1). 10.1371/journal.ppat.1001261 WOS:000286654100018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li G, Zhou X, Xu J-R. Genetic control of infection-related development in Magnaporthe oryzae. Current Opinion in Microbiology. 2012;15(6):678–84. 10.1016/j.mib.2012.09.004 WOS:000313612200007. [DOI] [PubMed] [Google Scholar]
- 50. Lee YH, Dean RA. cAMP regulates infection structure formation in the plant-pathogenic fungus Magnaporthe grisea . Plant Cell. 1993;5(6):693–700. 10.1105/tpc.5.6.693 WOS:A1993LH89900011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Verstrepen KJ, Klis FM. Flocculation, adhesion and biofilm formation in yeasts. Molecular Microbiology. 2006;60(1):5–15. 10.1111/j.1365-2958.2006.05072.x WOS:000235893500002. [DOI] [PubMed] [Google Scholar]
- 52. Wang G, Li G, Zhang S, Jiang C, Qin J, Xu J-R. Activation of the signalling mucin MoMsb2 and its functional relationship with Cbp1 in Magnaporthe oryzae. Environmental Microbiology. 2015;17(8):2969–81. 10.1111/1462-2920.12847 WOS:000360048800030. [DOI] [PubMed] [Google Scholar]
- 53. Talbot NJ, Kershaw MJ, Wakley GE, deVries OMH, Wessels JGH, Hamer JE. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell. 1996;8(6):985–99. WOS:A1996UV19500005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. DeZwaan TM, Carroll AM, Valent B, Sweigard JA. Magnaporthe grisea Pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11(10):2013–30. 10.1105/tpc.11.10.2013 WOS:000083430200018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Franck WL, Gokce E, Oh Y, Muddiman DC, Dean RA. Temporal Analysis of the Magnaporthe Oryzae Proteome During Conidial Germination and Cyclic AMP (cAMP)-mediated Appressorium Formation. Molecular & Cellular Proteomics. 2013;12(8):2249–65. 10.1074/mcp.M112.025874 WOS:000328839100017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Irie T, Matsumura H, Terauchi R, Saitoh H. Serial Analysis of Gene Expression (SAGE) of Magnaporthe grisea: genes involved in appressorium formation. Molecular Genetics and Genomics. 2003;270(2):181–9. 10.1007/s00438-003-0911-6 WOS:000186542500010. [DOI] [PubMed] [Google Scholar]
- 57. Yu D, Ranathunge K, Huang H, Pei Z, Franke R, Schreiber L, et al. Wax Crystal-Sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta. 2008;228(4):675–85. 10.1007/s00425-008-0770-9 WOS:000259008800015. [DOI] [PubMed] [Google Scholar]
- 58. Gravelat FN, Beauvais A, Liu H, Lee MJ, Snarr BD, Chen D, et al. Aspergillus Galactosaminogalactan Mediates Adherence to Host Constituents and Conceals Hyphal beta-Glucan from the Immune System. Plos Pathogens. 2013;9(8). 10.1371/journal.ppat.1003575 WOS:000323888200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Beaussart A, El-Kirat-Chatel S, Fontaine T, Latge J-P, Dufrene YF. Nanoscale biophysical properties of the cell surface galactosaminogalactan from the fungal pathogen Aspergillus fumigatus. Nanoscale. 2015;7(36):14996–5004. 10.1039/c5nr04399a WOS:000360831100024. [DOI] [PubMed] [Google Scholar]
- 60. Mysyakina IS, Bokareva DA, Usov AI, Feofilova EP. Differences in the carbohydrate composition between the yeastlike and mycelial cells of Mucor hiemalis. Microbiology. 2012;81(4):405–8. 10.1134/s0026261712040133 WOS:000307345900004. [DOI] [PubMed] [Google Scholar]
- 61. Cantu D, Greve LC, Labavitch JM, Powell ALT. Characterization of the cell wall of the ubiquitous plant pathogen Botrytis cinerea. Mycological Research. 2009;113:1396–403. 10.1016/j.mycres.2009.09.006 WOS:000273239300006. [DOI] [PubMed] [Google Scholar]
- 62. Ahrazem O, Prieto A, Gimenez-Abian MI, Leal JA, Jimenez-Barbero J, Bernabe M. Structural elucidation of fungal polysaccharides isolated from the cell wall of Plectosphaerella cucumerina and Verticillium spp. Carbohydrate Research. 2006;341(2):246–52. 10.1016/j.carres.2005.10.021 WOS:000234869700008. [DOI] [PubMed] [Google Scholar]
- 63. Schoffelmeer EAM, Klis FM, Sietsma JH, Cornelissen BJC. The cell wall of Fusarium oxysporum. Fungal Genetics and Biology. 1999;27(2–3):275–82. 10.1006/fgbi.1999.1153 WOS:000082047900019. [DOI] [PubMed] [Google Scholar]
- 64. Shibata N, Ikuta K, Imai T, Satoh Y, Satoh R, Suzuki A, et al. Existence of branched side-chains in the cell wall mannan of pathogenic yeast, Candida albicans—Structure-antigenicity relationhip between the cell wall mannans of Candida albicans and Candida parapsilosis . Journal of Biological Chemistry. 1995;270(3):1113–22. WOS:A1995QB15600024. [DOI] [PubMed] [Google Scholar]
- 65. Thieme TR, Ballou CE. Nature of phosphodiester linkage of phosphomannan from yeast Koleckera brevis . Biochemistry. 1971;10(22):4121-& 10.1021/bi00798a017 WOS:A1971K639500018. [DOI] [PubMed] [Google Scholar]
- 66. Cawley TN, Letters R, Harringt. Mg. Study of phosphate linkages in phosphomannan in cell walls of Saccharomyces cerevisiae . Biochemical Journal. 1972;129(3):711-& WOS:A1972N574600020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cabib E, Farkas V, Kosik O, Blanco N, Arroyo J, McPhie P. Assembly of the Yeast Cell Wall: Crh1p and Crh2p act as transglycosylases in vivo and in vitro . Journal of Biological Chemistry. 2008;283(44):29859–72. 10.1074/jbc.M804274200 WOS:000260366900030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gilbert NM, Baker LG, Specht CA, Lodge JK. A Glycosylphosphatidylinositol Anchor Is Required for Membrane Localization but Dispensable for Cell Wall Association of Chitin Deacetylase 2 in Cryptococcus neoformans. Mbio. 2012;3(1). 10.1128/mBio.00007-12 WOS:000303331400023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Odenbach D, Thines E, Anke H, Foster AJ. The Magnaporthe grisea class VII chitin synthase is required for normal appressorial development and function. Molecular Plant Pathology. 2009;10(1):81–94. 10.1111/j.1364-3703.2008.00515.x WOS:000262151400008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9). 10.1093/nar/29.9.e45 WOS:000168605800024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sweigard J, Chumley F, Carroll A, Farrall L, Valent B. A series of vectors for fungal transformation. Fungal Genetics Newsletter 1997. p. 52–3. [Google Scholar]
- 72. Samalova M, Johnson J, Illes M, Kelly S, Fricker M, Gurr S. Nitric oxide generated by the rice blast fungus Magnaporthe oryzae drives plant infection. New Phytologist. 2013;197(1):207–22. 10.1111/j.1469-8137.2012.04368.x WOS:000311606800023. [DOI] [PubMed] [Google Scholar]
- 73. Oldenburg KR, Vo KT, Michaelis S, Paddon C. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Research. 1997;25(2):451–2. 10.1093/nar/25.2.451 WOS:A1997WD44400025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, et al. Short protocols in Molecular Biology, 4th Edn New York: Wiley; 1999. [Google Scholar]
- 75. Samalova M, Meyer AJ, Gurr SJ, Fricker MD. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytologist. 2014;201(2):556–73. 10.1111/nph.12530 WOS:000328511700020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Conserved active site residues are marked in green boxes, and the zinc binding triad in blue boxes.
(TIF)
A) Development of appressoria (black arrow head) at 5 hpi on rice leaves. B) Melanized appressoria (white arrow head) and invasive hyphae (black arrow head) at 36 hpi on rice leaves. Scale bars: 20 μm. C) Expression of CDAs during in planta development at 36 hpi. Error bars show standard deviation, n = 3.
(TIF)
A) Schematic of deletion strategy, whereby the gene of interest is replaced by a gene encoding antibiotic resistance. Arrows show position of primers used to screen transformants. Pair 1 amplifies the gene of interest, confirming its absence in the deletion strain. Pairs 2 and 3 test for integration of the deletion construct at the desired locus. B) PCR analysis of single CDA deletion strains, showing successful deletion of CBP1, CDA2 and CDA3. C) PCR analysis of double CDA deletion strains (gene tested for written in red). D) PCR analysis of triple CDA deletion strain, showing successful deletion of CDA3. Note: Some of the gel pictures are composites images of different parts of the same gel.
(TIF)
A) Targeted gene deletion strategy, whereby the gene of interest is replaced by a gene encoding resistance to an antibiotic, due to homologous recombination between sequences (green). Southern blots showing successful single B), double C) and triple D) knockouts of chitin deacetylases.
Blots containing restriction digested gDNA of putative deletion strains were hybridised with α-32P labelled DNA homologous to the hygromycin, bialaphos or sulphonylurea resistance genes respectively. The cartoon above each blot shows the expected band size based upon the position of the restriction enzyme sites at each locus. Successful gene replacement is evidenced by a single band of the expected size, (as calibrated against standard gel size marker ladder and with band sizes given in kilobases (kb). For the double deletion strains, cross-hybridisation was observed between the hygromycin and bialaphos resistance cassettes, due to a common promoter sequence. These faint bands are labelled in B.
(TIF)
Conidia of the WT and cda2/cbp1/cda3 strains were inoculated onto the surfaces in 50 μl droplets, and incubated for 24 hr. Scale bars: 40 μm.
(TIF)
A) OGA488 staining of WT, cda2/cbp1 and cda2/cbp1/cda3 germlings at 24 hpi, showing absence of labelling in appressoria of the deletion strains (white arrow heads). B) Calcofluor white staining of germlings at 16 hpi, showing abnormally elongated, septate germ tubes (white arrow heads) in mutant strains, and abnormally shaped appressoria in cbp1. Scale bars: 20 μm.
(TIF)
A) Cbp1:mCherry and B) Cda2:mCherry fluorescence at later stages of appressorium development (as indicated). Scale bars: 10 μm.
(TIF)
Molecular structure, degree of polymerization and degree of deacetylation (DDA) are indicated (when known).
(TIF)
A) Pathogenicity of the cda2/cbp1/cda3 deletion strain on rice leaves, showing comparable lesion density to the WT strain (two independent triple deletion lines are shown). A mock inoculation of 0.2% gelatine was included as a negative control. Scale bars: 1 cm. B) Quantification of lesion numbers on detached rice leaves inoculated with different cda mutant strains, normalized to the Guy11 WT strain (± SD, n = 3).
(TIF)
Appressorium development was observed in cda2/cbp1/cda3 (white arrow heads) in the presence of 1-octacosanol, but germ tubes remained abnormally elongated. Scale bars: 50 μm.
(TIF)
A) Germlings of the WT and cda2/cbp1/cda3 strain, stained with FITC-ConA. Pictures were taken from Experiment 1 (quantified in B). Scale bars: 50μm. B) Fluorescence intensity of germlings stained with FITC-ConA. Fluorescence intensities of ~140 germ tubes were measured, across 4 independent experiments. White bars: WT, grey bars: cda2/cbp1/cda3. Significant differences were found in 3 of the 4 experiments (2-way ANOVA with post-hoc Tukey test (** = p <0.01, *** = p < 0.001))
(TIF)
(TIF)
(XLS)
(XLS)
Conidia of the WT and cda mutants were inoculated onto an artificial surface and incubated for 16 hr, in the presence of IBMX (3-isobutyl-1-methylxanthine), HDD (1,16 hexadecanediol; cutin monomer) or DAG (1,2-dioctanoyl-sn-glycerol). Significant (Mann-Whitney U-test, p < 0.001) reductions (asterisks) in germ tube lengths were observed in the deletion strains with all treatments, when compared with the control. ND—Not determined.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.