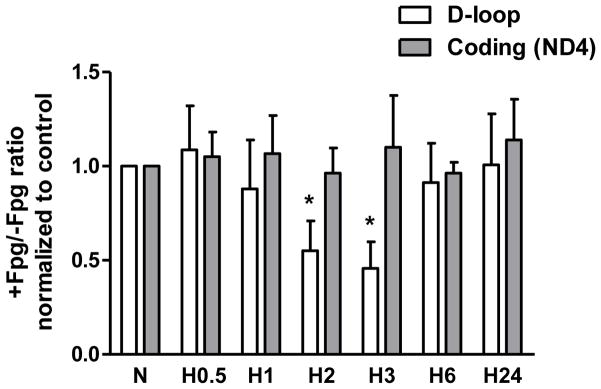
Fig. 3.
Results of Fpg-sensitive quantitative real time PCR analysis of oxidative DNA damage in D-loop and coding regions of mtDNA from normoxic (N) rat pulmonary artery endothelial cells and cells exposed to hypoxia for 0.5, 1, 2, 3, 6 and 24 hours (H0.5 – H24). The ratio of PCR product accumulation in treated and untreated with Fpg samples is indicative of oxidative DNA damage. Mean ± SE, N = 3, *P < 0.05, significantly different from normoxic controls.