Figure 5. Application of SLENDR by Multiplex Genome Editing.
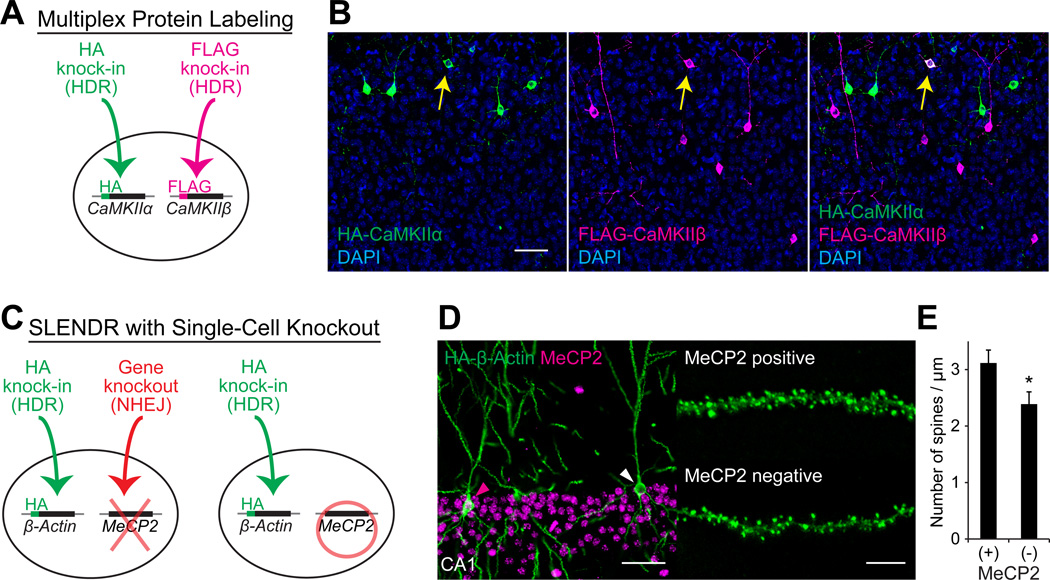
(A) Schematics of multiplex labeling of different endogenous proteins with different tags. The HA and FLAG sequences are inserted to CaMKIIα and CaMKIIβ, respectively, in the same cell through HDR-mediated genome editing.
(B) Multiplex labeling of endogenous CaMKIIα and CaMKIIβ. Confocal microscopic images of the cerebral cortex at P14 showing the DAPI signal (blue) and immunoreactivities for the HA tag (green) and the FLAG tag (magenta) fused to the N-terminus of endogenous CaMKIIα and CaMKIIβ, respectively. The yellow arrows indicate HA and FLAG double-positive layer 2/3 neurons.
(C) Schematics of combining SLENDR with NHEJ-mediated gene knockout. The HA sequence is inserted to β-Actin through HDR-mediated genome editing and a frame-shift mutation is induced in MeCP2 through NHEJ-mediated genome editing in the same cell (left). In our strategy, some cells in the same tissue undergo only HDR-mediated genome editing (right), allowing comparison of the expression and localization of endogenous proteins within the same brain slice.
(D) Immunofluorescence images of MeCP2 (magenta) and HA-β-Actin (green) in the hippocampus CA1 region. Magenta arrowhead, MeCP2-positive; white arrowhead, MeCP2-negative. Representative images of secondary dendrites of MeCP2-positive and negative neurons (right).
(E) The averaged density of spines on secondary or tertiary apical dendrites in MeCP2-positive (n = 527 spines/ 6 neurons) and negative (n = 437/5) neurons. *p < 0.05, Student’s t test.
Data are represented as mean ± SEM.
Scale bars, 50 µm (B and D, left); 5 µm (D, right).
See also Table S3.