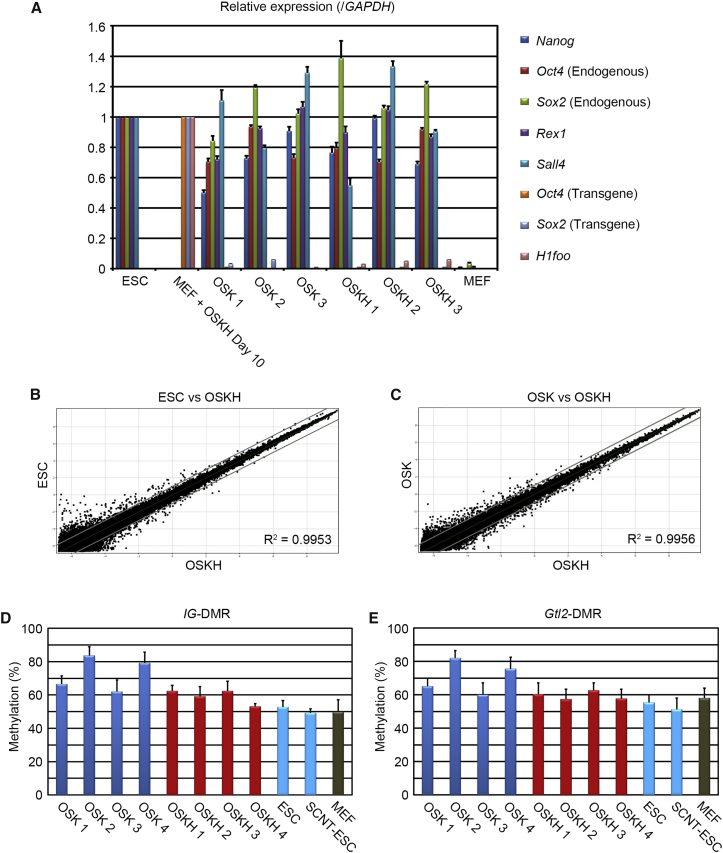
Figure 2.
Characteristics of OSKH-Induced iPSCs
(A) qRT-PCR analysis of pluripotency genes and H1foo in iPSCs. Error bars represent the SEM (n = 3 independent experiments).
(B and C) Pairwise scatterplots of global gene-expression cDNA microarray patterns of OSKH-iPSCs (n = 4) compared with ESC (n = 1) (B) or OSK-iPSCs (n = 3) (C). The gray lines indicate log2 2-fold changes in gene-expression levels between the paired cell types.
(D and E) Degree of DNA methylation at IG-DMR and Gtl2-DMR in four OSK iPSC clones, four OSKH-iPSC clones, and ESCs, as well as MEFs analyzed by pyrosequencing. Error bars represent the SEM (n = 3 independent experiments).