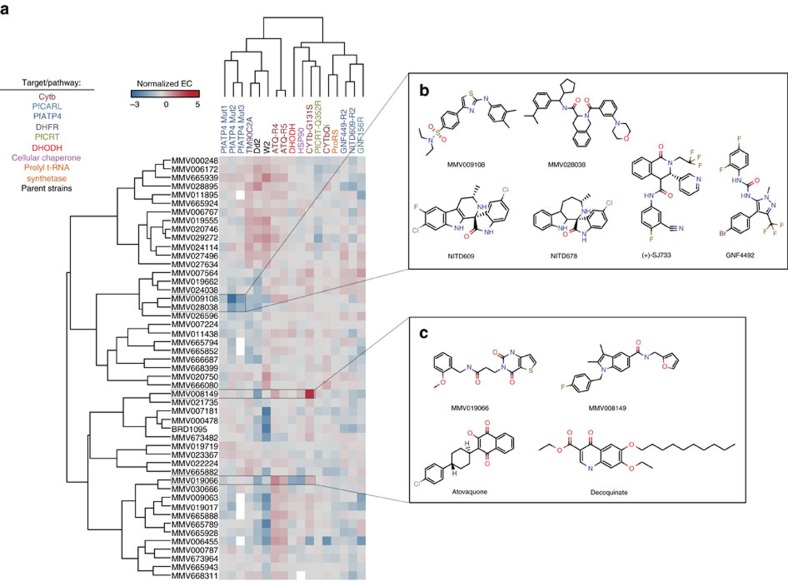
Figure 2. Cross-resistance fold shifts observed in compound set.
(a) A total of 15 resistant strains were tested with each MMV compound to identify potential pre-existing cross-resistance. Calculating the fold shifts between each clone and either a corresponding parent or a drug sensitive 3D7 strain generated the heatmap. To normalize conferred resistance and sensitivity, the natural log of each fold shift is displayed. Fold shifts instead of raw data were used as multiple assays were run with varying times and detection indicators. Incomplete cross-resistance assays are depicted in white. All assays were run in triplicate. For one compound (BRD1095), a close analogue (BRD3444) was analysed for TM90C2A and PfATP4-Mut1–3. SMILEs for all compounds are listed in Supplementary Data 1. (b) Chemical structures of the two MMV compounds (MMV009108 and MMV028038) with increased efficacy against one or more pfatp4 mutated clones. Both compounds displayed low structural similarity to a number of other known pfatp4 inhibitors. (c) Chemical structures of the two MMV compounds (MMV019066 and MMV008149) with decreased efficacy against one or more cytochrome bc1 mutated clones. Atovaquone and decoquinate, two other cytochrome bc1 inhibitors, were structurally significantly different.