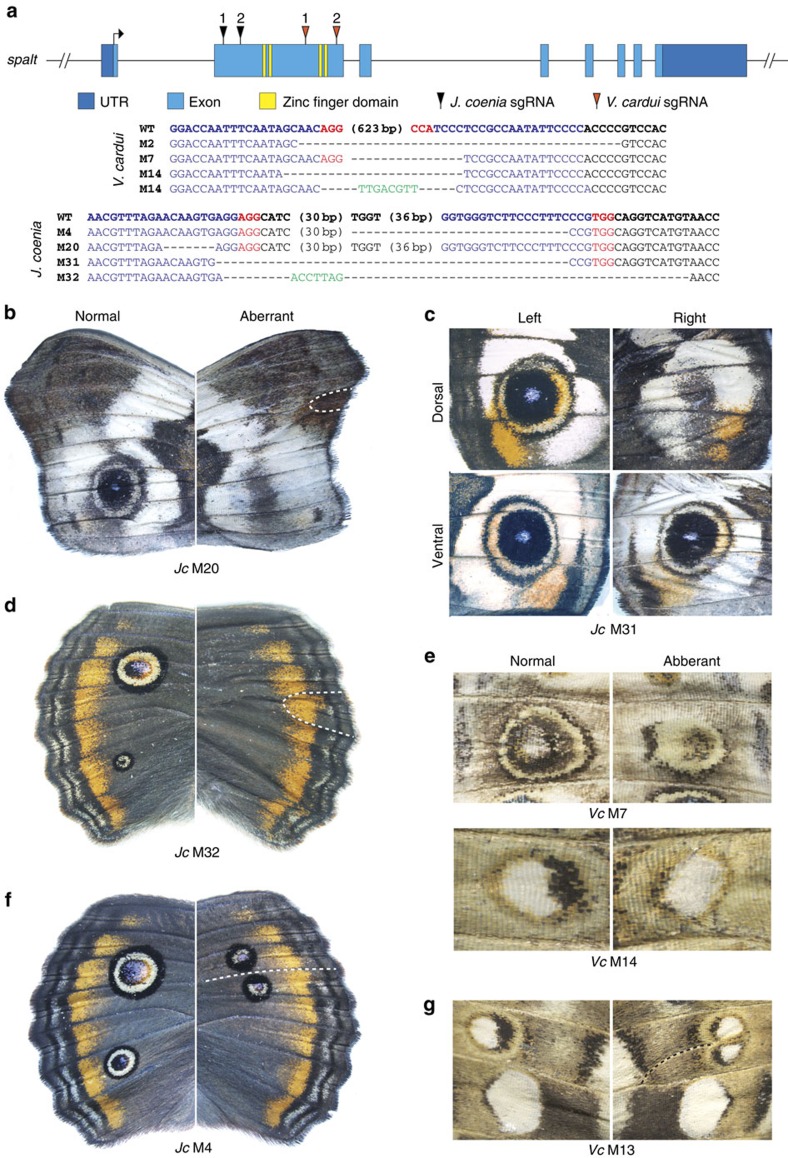
Figure 2. CRISPR/Cas9 spalt deletions result in reduction and loss of eyespot colour patterns.
(a) Location of sgRNAs relative to the V. cardui spalt locus. Sequences of spalt alleles from the animals shown confirm lesions in target regions. Blue: sgRNA targets. Red: PAM sequences. Green: novel sequences not observed in wild-type alleles. Somatic deletions in spalt cause complete loss of forewing eyespots in J. coenia: (b) ventral forewing; (c) dorsal forewing; (d) dorsal hindwing. (e) spalt deletions reduce eyespots in V. cardui ventral hindwings (top) and forewings (bottom). Ectopic wing veins (dashed lines) resulting from spalt deletion subdivide eyespot patterns in (f) J. coenia (also showing a missing posterior eyespot), and (g) V. cardui. All comparisons shown are left–right asymmetrical phenotypes from individual injected butterflies. ‘Normal' phenotypes are as wild-type, ‘aberrant' phenotypes are non-wild type and vary asymmetrically from the normal patterns on the same animal.