Fig. 3.
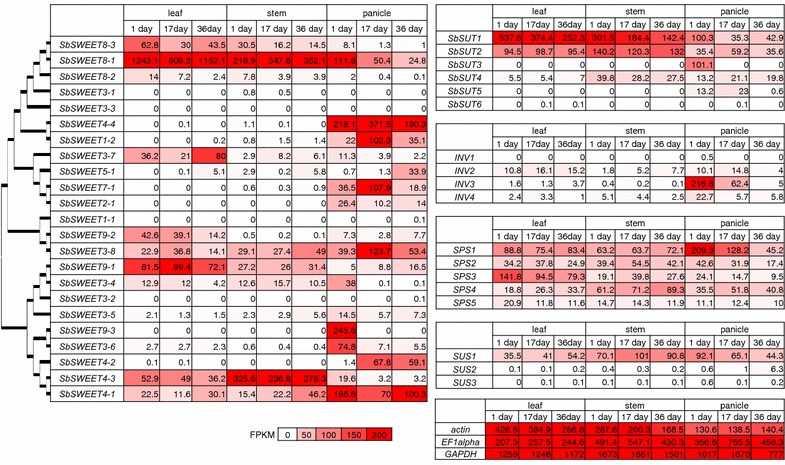
FPKM values of sugar-related genes at the sucrose accumulation stage. FPKM (fragments per kilobase of exon per million mapped sequence reads) values reflect the quantities of existing RNA of each paralog in the cells or tissues. FPKM values for SWEET, SUT, INV, SPS, SUS, actin, elongation factor 1-alpha (EF1alpha), and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) are shown as heatmaps. Actin, EF1alpha, and GAPDH are constitutively expressed controls. The number in each box indicates the FPKM value of each gene. Boxes at the bottom indicate the reference color intensities of FPKM values. Samples were extracted from the leaf, stem, or panicle on days 1, 17, and 36 after heading (the stage of sucrose accumulation in the stem). Phylogenetic trees of 23 putative sorghum SWEET genes are also shown on the left side
