Fig. 1.
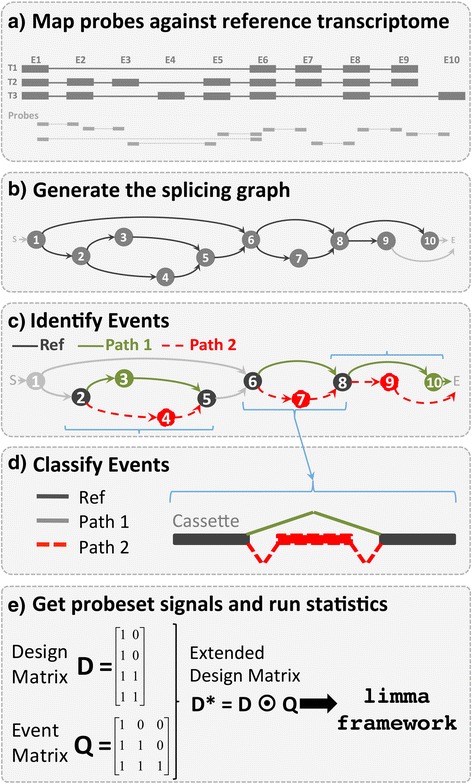
Overview of EventPointer. a Using as input a reference transcriptome (Ensembl), the probes of the array are mapped against it, b the splicing graph is created. c Using the splicing graph, EventPointer detects every possible event by defining common nodes and edges (Reference Path) and two alternative paths (P1 and P2). d These events are classified into the canonical splicing categories. e Finally, for a specific experiment and using the design matrix of the experiment and an auxiliary event matrix, the statistical significance of each gene is computed using the limma framework. Steps (a-d) are specific of the array and thus, is only necessary to rerun them if the reference transcriptome is changed. Step E) must be performed for each experiment
