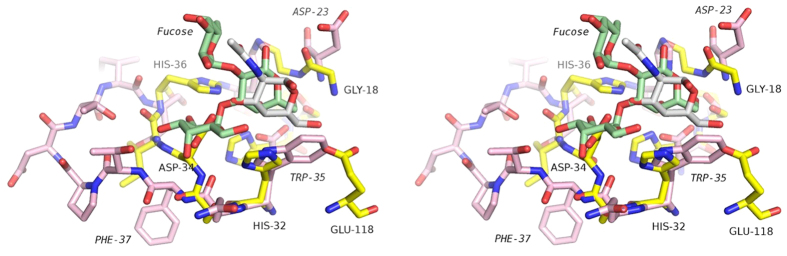
Figure 5. An overlap of liganded MOA and Mytilec monomers.
Superposition was carried out with SSM27. The program selected 130 residues for overlap, with only 9.2% sequence identity, giving an rmsd of 1.7 Å. The carbon atoms of MytiLec and the GalNAc ligand are shown in yellow and white respectively. MOA and the blood group B trisaccharide, Galα(1,3) [Fucα(1,2)]Gal, are coloured with pink and pale green carbon atoms respectively17. The figure shows a close-up of site 1. Residue numbers for MOA are shown in an italic font. It can be seen that the non-reducing galactose residue of the trisaccharide overlaps the GalNAc bound to MytiLec, but Asp 34 of MytiLec clashes severely with the reducing Gal residue. Equivalent clashes are seen at each MytiLec binding site. Overall the binding sites of the two proteins are very different, with MOA having a much more extended surface loop, and Trp 35 (MOA) replacing His 32 (MytiLec). Behind Trp 35, it can be seen that His 15 of MytiLec is replaced by Asp 20 in MOA.