FIGURE 3.
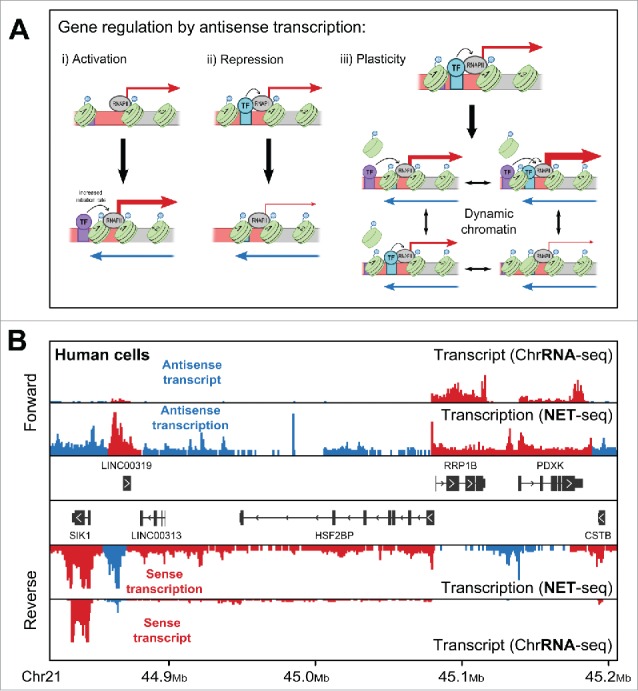
(A) Three, mutually inclusive models for how antisense transcription might regulate genes. i) A gene with a single, nucleosome-occluded transcription factor (TF) binding site (purple box) in its promoter shows a moderate level of sense transcription. In the presence of antisense transcription (blue arrow), chromatin remodeling leads to the movement of this nucleosome, resulting in increased sense transcription (indicated by the larger red arrow). In ii) the situation is reversed, with antisense transcription leading to occlusion of a different binding site (turquoise box), causing reduced sense transcription. iii) Here, antisense transcription sets up a dynamic chromatin at the promoter allowing switching between different chromatin states, each with a distinct sense transcription output. In a population of cells this would result in noisier gene expression. (B) A view of transcription of both strands (forward and reverse) of a region of chromosome 21 in human HeLa cells, analogous to Figure 1B. Shown are the levels of transcript, measured using ChrRNA-seq (a modification of RNA-seq that aims to measure both processed and unprocessed transcript levels), and transcription, using NET-seq.32 Reads have been colored as reflecting either sense (red) or antisense (blue) transcription/transcript based on the orientation of their nearest gene.
