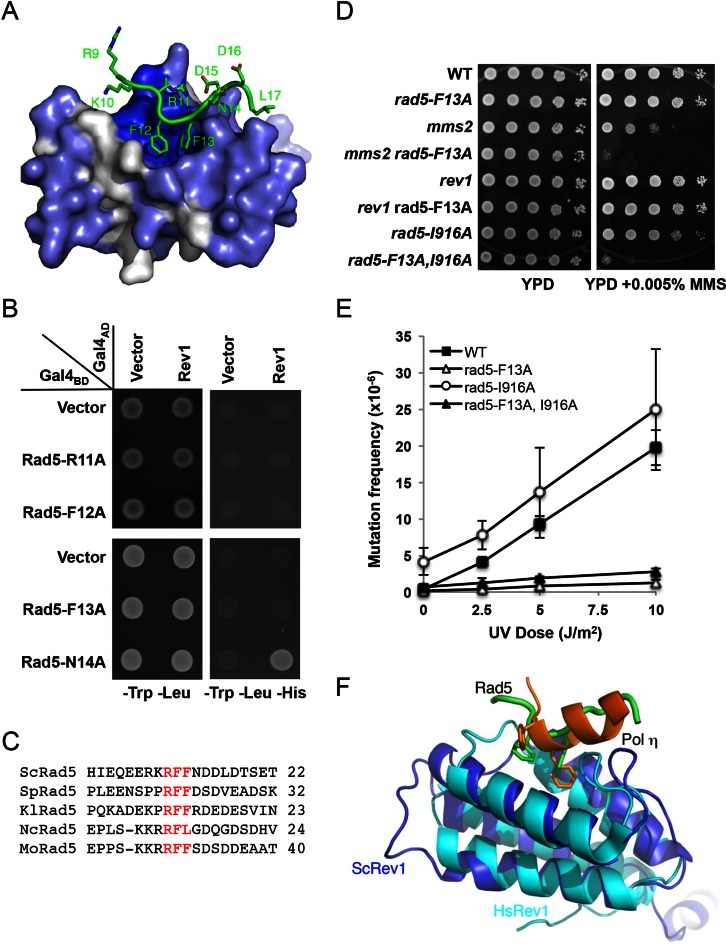
Figure 7.
Characterization of the Rad5-NTD RFF motif. (A) Rad5-NTD residues involved in binding to a conserved pocket of Rev1. Rev1 is shown as gray surface with conserved residues in blue and the Rad5 polypeptide is shown in green. (B) The ‘RFF’ motif of Rad5 is essential for the binding of Rad5 to Rev1, as judged by a Y2H assay. The experimental conditions were as described in Figure 1. (C) Sequence alignments around the ‘RFF’ motif among several fungal Rad5 homologs. The RFF motif is in red. Source of sequences: ScRad5, Saccharomyces cerevisiae, P32849.1; SpRad5, Schizosaccharomyces pombe, XP_001713034.1; KlRad5, Kluyveromyces lactis, XP_455865.1; NcRad5, Neurospora crassa, XP_958511.1; MoRad5, Magnaporthe oryzae, XP_003712540.1. (D) Inter- and intragenic interactions between rad5-F13A and DDT pathways mutations with respect to MMS-induced killing. (E) Effects of rad5-F13A and rad5-I916A on UV-induced mutagenesis. Strains are isogenic derivatives of DBY747. Data are the average of at least three independent experiments with standard deviation. (F) Structural alignment of Rev1 between budding yeast (blue) and human (cyan, PDB code 2LSK). hPolη (orange) binds to the same pocket of hRev1 as yRad5 (green) does with yRev1.