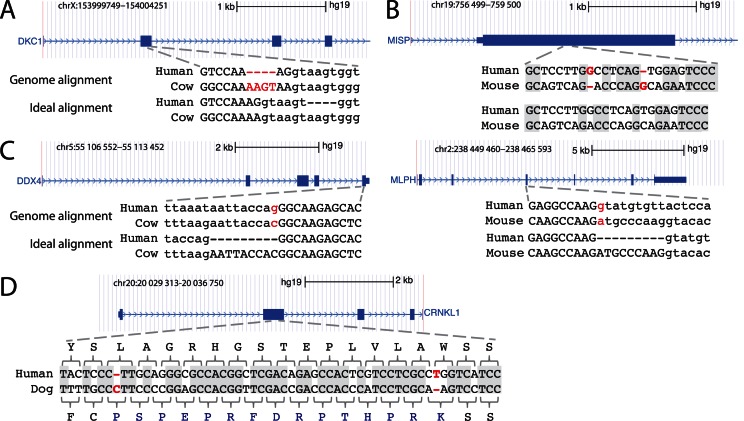
Figure 1.
Limitations of genome alignments for assessing exon conservation. (A) The genome alignment shows a 4 bp frameshifting insertion (red font). This is an alignment ambiguity, as an equivalent alignment exists where this insertion is in the intron (‘ideal alignment’). Upper case letters are exonic bases, lower case letters are intronic bases. (B) The genome alignment shows two close frameshifts that compensate each other. These two frameshifts are likely spurious and did not happen in evolution as an alternative alignment with 12 versus 13 identical bases (grey background) exist that lacks these indels. (C) Two examples where the acceptor (left) or donor (right) splice site is mutated. In both cases, the exon is conserved but its splice site has shifted by 9 bp into the intron. The ideal alignment would align the original and the shifted splice site. By aligning non-orthologous but ‘functionally equivalent’ splice site bases, the ideal alignment correctly identifies the exon boundaries in the other species. (D) In contrast to (B), two real compensating frameshifts change the reading frame for 15 codons. The alignment with both frameshifts has a much higher number of identical bases (grey background) than the alignment without both frameshifts, which strongly suggests that these compensating frameshifts did occur in evolution.