Abstract
Detection of cell-free DNA in liquid biopsies offers great potential for use in non-invasive prenatal testing and as a cancer biomarker. Fetal and tumor DNA fractions however can be extremely low in these samples and ultra-sensitive methods are required for their detection. Here, we report an extremely simple and fast method for introduction of barcodes into DNA libraries made from 5 ng of DNA. Barcoded adapter primers are designed with an oligonucleotide hairpin structure to protect the molecular barcodes during the first rounds of polymerase chain reaction (PCR) and prevent them from participating in mis-priming events. Our approach enables high-level multiplexing and next-generation sequencing library construction with flexible library content. We show that uniform libraries of 1-, 5-, 13- and 31-plex can be generated. Utilizing the barcodes to generate consensus reads for each original DNA molecule reduces background sequencing noise and allows detection of variant alleles below 0.1% frequency in clonal cell line DNA and in cell-free plasma DNA. Thus, our approach bridges the gap between the highly sensitive but specific capabilities of digital PCR, which only allows a limited number of variants to be analyzed, with the broad target capability of next-generation sequencing which traditionally lacks the sensitivity to detect rare variants.
INTRODUCTION
The ability of massively-parallel, next-generation DNA sequencing (NGS) to identify low prevalence mutations in heterogeneous samples has revolutionized basic and translational research in cancer and many other fields (1). However, detection of sequence variants below 1% frequency remains a challenge with standard NGS protocols due to background noise, much of which is introduced by polymerases during library construction (2). This background noise is problematic in many clinical and research applications, including detection of rare sequence variants in liquid biopsies for non-invasive prenatal diagnostics (NIPD) or for biomarker applications in cancer.
Detection and analysis of fetal DNA in maternal plasma has led to a revolution in NIPD for Downs Syndrome and other disorders involving large chromosomal abnormalities (3,4). Moving forward, detection of single nucleotide variants specific to the fetus offers the potential to diagnose monogenic disorders early on in pregnancy without the risks associated with chorionic villus sampling or amniocentesis (5–7). In cancer, applications of rare mutation detection in liquid biopsies include analysis of tumor heterogeneity and identification of therapy resistant clones(8), monitoring clonal evolution and response to therapy (9) and early cancer diagnosis using blood/plasma, sputum, urine or other bodily fluids (10–12). In many cases, these scenarios potentially require detection of variant allele fractions of 0.1% or less.
In both NIPD and cancer biomarker research, the introduction of COLD polymerase chain reaction (PCR) (13,14) more recently digital PCR (15) technologies has enabled detection and quantification of ultra-rare sequence variants in liquid biopsies (16,17). However, digital PCR assays are specific for both nucleotide position and the specific base change. Combined with the fact that multiplexing capability is limited, digital PCR is most useful in situations where a known variant is being sought or where disease-related variants are well characterized and limited in number. For recessive disorders, mutations in tumor suppressor genes and even recurrent mutations in many oncogenes, de novo detection of variants at many base positions is typically required and digital PCR is not the answer. Instead, sensitive sequencing approaches such as targeted deep sequencing, duplex sequencing or molecular barcoding offer an attractive alternative (18–22) although they typically require complex library construction protocols.
Introduction of molecular barcodes (random oligonucleotide sequences e.g. N12-14) to uniquely tag individual target DNA molecules can be used to identify and reduce sequencing errors introduced during NGS library construction (Supplementary Figure S1) and enables robust detection of ultra-rare variants (20,23). Ligation of barcodes onto target DNA followed by target capture and amplification is inefficient and risks missing rare variants when using low DNA inputs such as those obtained from liquid biopsies. Introduction of barcodes by PCR can be achieved with low DNA inputs (20) but the random barcode sequences behave promiscuously resulting in formation of non-specific PCR products. Consequently, multiplexing is challenging and library construction requires complex, multi-step workflows, some of which include gel purification of PCR products (20). Here, we report development of a library construction approach that uses reduced primer concentrations, elongated PCR extension times and hairpin-protected barcode primers to enable Simple, Multiplexed, PCR-based barcoding of DNA for Sensitive mutation detection using Sequencing (SiMSen-Seq). SiMSen-Seq facilitates detection of sequence variants at or below 0.1% allele frequency, works with low DNA input (<50 ng) and can be used to interrogate multiple genome loci covering >1 kb of target sequence if desired.
MATERIALS AND METHODS
DNA
Wild-type genomic DNA was extracted from a clonally derived Barrett's esophageal cell line, CP-A, using the QIAamp DNA Mini kit (Qiagen). Wild-type circulating, cell-free DNA (ccfDNA) was extracted from pooled patient plasma (Innovative Research) using QIAamp Circulating Nucleic Acid kit (Qiagen). DNA concentrations were quantified with the Qubit 2.0 Fluorometer (Life Technologies) and stored at −20°C. Genomic DNA was sheared using a M220 focused-ultrasonicator (Covaris).
Melting curve analysis
Hairpin stability was analyzed by melting curve analysis using Varian Cary 300 UV-Vis spectrophotometer (Varian, Inc). Primers were analyzed at a concentration of 1 μM in PCR buffer (10 mM Tris–HCl (pH 8.0), 50 mM KCl and 5 mM MgCl2). Samples were degased using preheating at 90°C for 10 min. The absorbance was measured at 260 nm with a temperature gradient from 25 to 90°C, increasing the temperature stepwise, 0.4°C/min. Data were recorded every 0.4°C.
Barcoding and library construction
Barcoding of DNA was performed with PCR in 10 μl using 1× AccuPrime PCR Buffer II, 0.2 U AccuPrime Taq DNA Polymerase High Fidelity (both Invitrogen, Thermo Fisher Scientific), 40 nM of each primer (IDT, Inc) and 5–100 ng DNA. Primer sequences are shown in Supplementary Table S1. The temperature profile was 98°C for 3 min followed by three cycles of amplification (98°C for 10 s, 62°C for 6 min and 72°C for 30 s), 65°C for 15 min and 95°C for 15 min. Twenty microliter TE buffer, pH 8.0 (Ambion, Thermo Fisher Scientific) with final concentration of 30 ng/μl protease (Streptomyces griseus, Sigma Aldrich) was added to inactivate the Taq DNA polymerase at the 65°C for 15 min step. The second round of PCR was performed in 40 μl using 1× Q5 Hot Start High-Fidelity Master Mix (New England BioLabs), 400 nM of each Illumina adaptor primer and 10 μl PCR products from the first round of PCR. The temperature profile was 95°C for 3 min followed by 18–30 cycles of amplification (98°C for 10 s, ramping from 80°C down to 72°C and up 76°C, 0.2°C per 1 s increments, 76°C for 30 s). Thirty-six microliter PCR products were purified using the Agencourt AMPure XP system (Beckman Coulter, Inc.) according to the manufacturers’ instructions. The applied volume ratio between beads and PCR products ranged from 0.83 to 1.0, depending on amplicon length. The purified product was eluted in 20 μl TE buffer, PH 8.0 and prior to sequencing, library products were assessed on a Fragment Analyzer (Advanced Analytic Technology, Inc) to ensure correct sizing.
Sequencing
The products from the second round of PCR contained Illumina sequencing adaptor sequences and indexes and were therefore sequencer-ready. To assess the amplification status of primer pairs in multiplexed SiMSen-Seq reactions, some libraries were initially sequenced at low depth using MiSeq instruments with the Nano Kit V2 in 1 × 150 mode (Illumina). For full sequencing runs, libraries were multiplexed per lane and sequenced on MiSeq or HiSeq2500 instruments (Illumina) in single or paired 150 bp mode.
Sequence analysis
FASTQ files were aligned to hg19 using bwa mem (0.7.12) with output bam files sorted by position and indexed using samtools (0.1.19). A custom pipeline was used to build consensus sequences as follows: the amplicons in each library were identified in bam files according to library plexity; for example, five target amplicons were identified in 5-plex experiments. Valid reads within each amplicon were identified as those which contained a barcode sequence in the correct orientation relative to the sequence of the targeting primer and hairpin stem. Remaining reads were grouped into families by amplicon and random 12mer barcode. For reads within each family, alignment information for individual reads was used to determine a consensus identity for bases (including indels) at each nucleotide position within the amplicons. This procedure is conceptually similar to that described in Schmitt et al. (19). Non-reference sequences were reported in consensus sequences if they composed 100% of the reads in families with 10–20 reads, or at least 90% of reads in families with >20 reads. The bioinformatics workflow is outlined in Supplementary Figure S2.
RESULTS
NGS library primers can be designed to be in open or closed configuration using a temperature-dependent hairpin structure
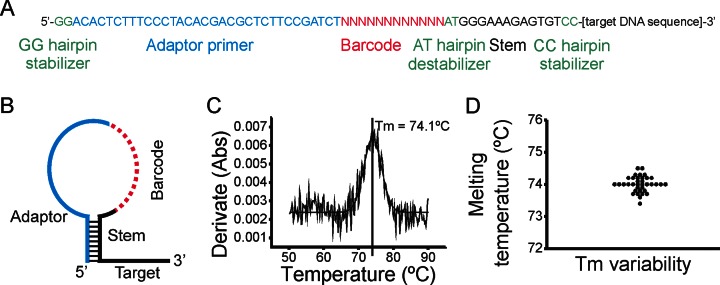
The major obstacle in PCR multiplexing is unwanted interactions between primers forming non-specific PCR products. The amount of non-specific PCR products depends on the number of primers multiplexed, but also on their length and sequence (24). Randomized sequences, such as barcodes, are potentially more prone to form non-specific PCR products, since they can interact promiscuously with adapter and target portions of all primers in the reaction. To help solve this issue, we designed a universal hairpin structure that protects the barcode and adapter sequences from spurious interaction, while leaving the target portion of the primer available for hybridization during the first steps of library construction (Figure 1A and B). While secondary structures such as panhandles and hairpins have been used to reduce primer-dimer (25) or to alter fluorescence characteristics of molecular probes (26), we believe that ours is the first use of a hairpin structure to protect a molecular barcode sequence during PCR. The hairpin protected barcode primer consists of: (i) standard target primer sequence, (ii) 12 randomized nucleotides used as barcode, (iii) adaptor primer sequence and (iv) 14 nucleotides forming a hairpin stem. The stem sequence was designed to be in a closed hairpin configuration at the PCR annealing temperature (60–62°C), but in an open state at the PCR elongation temperature (72–76°C). To minimize the primer length and hairpin size we used nucleotides in the sequencing adaptor region as a backbone to design the stem. Two additional guanine bases 5′ of the adaptor sequence (GG hairpin stabilizer) allowed us to increase the hairpin melting temperature. Furthermore, we included 2 nucleotides 3′ of the barcode (AT hairpin destabilizer) separating the barcode from the stem sequence. These nucleotides create two forced mismatches, ensuring that bases in the barcode do not strengthen the stem stability in a sub-fraction of the primers. To evaluate the hairpin melting temperature and its variability between primers we analyzed 36 primers with different DNA target sequences using melting curve analysis in a temperature controlled spectrophotometer (Figure 1C). All primers with the same hairpin-stem structure displayed almost identical melting temperature demonstrating a stable and robust hairpin design (mean ± SD = 74.0°C, ± 0.3°C; Figure 1D).
Figure 1.
SiMSen-Seq. (A) Sequence composition of hairpin protected barcode primer. Different sequence elements are indicated by color. (B) Schematic design and structure of hairpin protected barcode primer. (C) Melting curve analysis of hairpin protected barcode primer using a temperature controlled spectrophotometer. The derivative of the absorption over time is shown. The melting temperature (Tm) where 50% of primers are in an open configuration is indicated. (D). Thirty-six different hairpin protected barcode primers were evaluated (Mean = 70.01, SD ± 0.24).
Derivation of a simple, robust and fast library protocol using barcoding: SiMSen-Seq
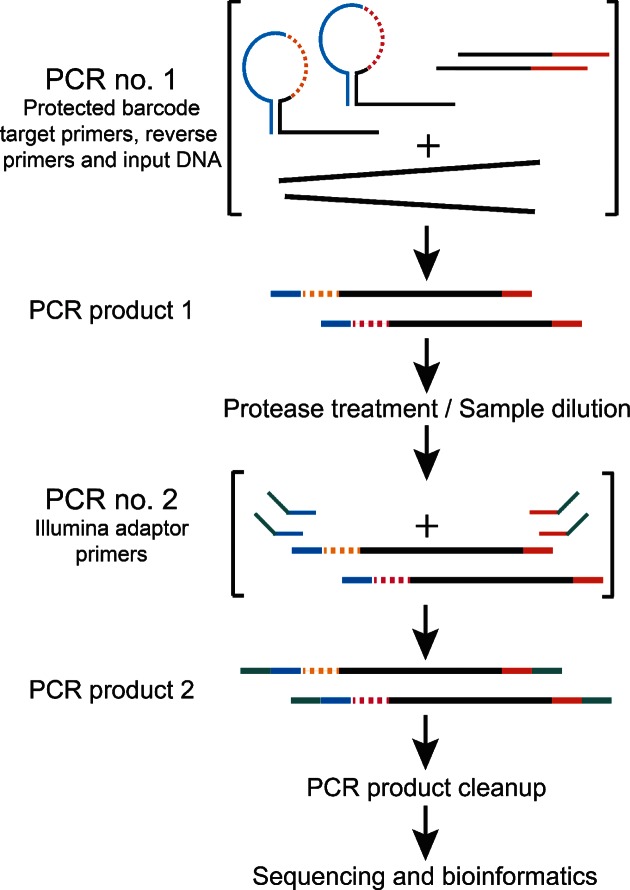
The SiMSen-Seq (Simple, Multiplexed, PCR-based barcoding of DNA for Sensitive mutation detection using Sequencing) approach essentially consists of two rounds of PCR using high fidelity DNA polymerases (Figure 2). In the first PCR, each target DNA is barcoded using the hairpin-protected barcode primers. To further reduce the formation of non-specific PCR products in the first PCR, we applied a standard multiplex pre-amplification strategy using 40 nM primer concentrations (10–20 times lower than in a standard PCR) and to compensate, the annealing time was extended to 6 min. The reaction was then terminated using a combined dilution and protease treatment step at 65°C for 15 min, to minimize the formation of non-specific PCR products in downstream handling. The resulting products were used directly in the second PCR step in which barcoded DNA molecules were amplified with Illumina adaptor primers to generate complete libraries. A PCR product clean-up was then performed with the Agencourt AMPure XP magnetic bead system.
Figure 2.
Schematic library construction workflow. In the first PCR consisting of three cycles, target DNA is amplified with hairpin protected barcode primers. The reaction is terminated with an incubation step that is a combined dilution and protease treatment step. In the second PCR that consists of 18–30 cycles, all individual amplicons are amplified to generate PCR products with Illumina adapter primers. Final libraries are purified with magnetic beads, normalized for concentration differences between samples and sequenced.
To test the effect of the hairpins on specific PCR product formation we evaluated 13 assays in both singleplex and multiplex library construction using primers with and without hairpins. For singleplex assays, fragment analysis data indicated an average increase in specific PCR product relative to non-specific of 1.73-fold when using hairpins (P < 0.01, Supplementary Figure S3A). For the 13-plex analysis, sequencing of unpurified libraries also indicated an average 1.76 fold increases in on-target yield when using the hairpin primers (P < 0.01, Supplementary Figure S3B and C).
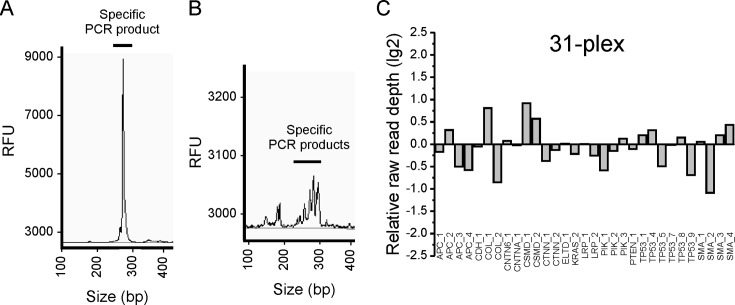
Using SiMSen-Seq we have successfully generated libraries targeting from 1 up to 31 different genomic DNA sequences in a single reaction (Figure 3A and B). Relative raw read uniformity between individual amplicons was evaluated for 5-, 13- and 31-plex libraries (Supplementary Figure S4 and Figure 3C). The relative read depth for each amplicon was within 1.3-fold of the mean with high reproducibility (SD < 0.12; n = 12) for the 5-plex libraries and within 2.1-fold of the mean for the 13-plex libraries (SD < 0.32: n = 3). The 31-plex library was sequenced only once and representation of all amplicons was within 1.9-fold of the mean. All SiMSen-Seq primers and tested multiplex combinations are shown in Supplementary Table S1.
Figure 3.
Library purity and uniformity. (A) Electropherogram of a purified final library targeting one DNA sequence using the Fragment Analyzer. (B) Electropherogram of a purified final library targeting 31 DNA sequences using the Fragment Analyzer. (C) Relative raw read depth of 31 multiplexed amplicons were analyzed. DNA from tumor cell line CP-A was used. The average raw read depth was 1.4 × 104 per amplicon.
SiMSen-Seq reduces sequencing errors of all nucleotides
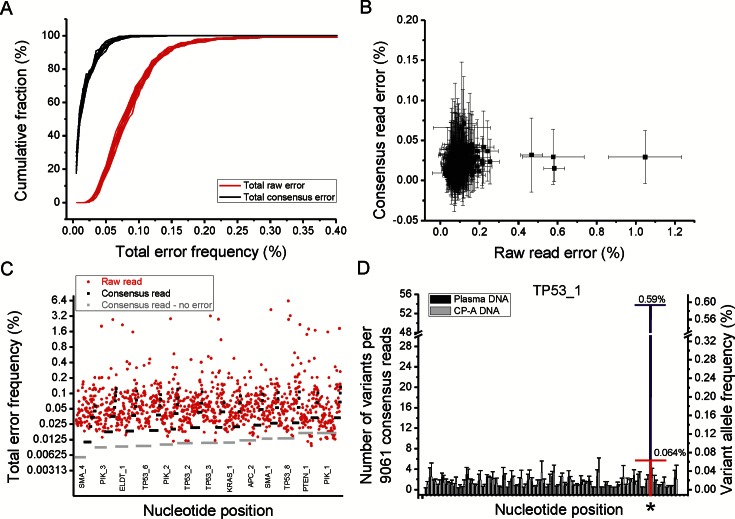
For sequencing error correction using SiMSen-Seq, raw reads mapping to the same amplicon position, and with the same unique barcode, were grouped into barcode families. Barcode families containing a minimum of 10 or 30 raw reads (depending on raw read depth) were then used to compute consensus reads. Consensus was determined for each base in the amplicons and we required 100% identical reads for families with 10–20 reads and ≥ 90% identical reads for families with > 20 reads. Figure 4A shows the uniform reduction of total error frequency using barcoding for 417 nucleotides across five amplicons analyzed in 12 replicates. The average error correction using consensus reads was 7.3-fold and the maximum correction for any nucleotide was 135-fold. About 40.2% of all nucleotides (2014 out of 5004 nucleotides) displayed no consensus read error and 99.3% of nucleotides showed a consensus error < 0.1% with 95% confidence (Figure 4B and Supplementary Figure S5). Four hot spot nucleotides (0.96% of all nucleotides) with raw read errors > 0.4% were identified and all were corrected to <0.05% error with barcoding. Next, we increased the multiplexing to 13 amplicons that covered 1042 nucleotides (Figure 4C). Data were consistent with the 5-plex experiment. Here, the average error correction was 7.2-fold, 59.5% of all nucleotides showed no consensus read error and 98.9% of all nucleotides showed a consensus error < 0.1%. Thirty nucleotides (30/1042; 2.9%) were hot spot positions for raw sequencing error (Supplementary Figure S6) and all were corrected to <0.07% error with barcoding (maximum correction factor was 475-fold). The five multiplexed amplicons analyzed above were also included in the 13-plex analysis and once again, four hot spot nucleotides were observed. However, only one hot spot nucleotide was common to both runs, while seven hot spot positions were different in the two experimental setups. This data also serves to illustrate that barcoding can eliminate sequencing errors that occur even with extremely deep sequencing (minimum read depth at hotspots in our study was 5.5 × 105). In all of the above analyses, DNA from the same clonally derived cell line, CP-A, were used. In all experiments, consensus read error was <0.15% for all base positions.
Figure 4.
SiMSen-Seq reduces PCR induced errors and enables rare mutant molecule detection. (A) Cumulative plot of total raw and consensus read errors. Total and consensus read error was calculated for of each of 417 base positions. Error frequency corresponds to the number of non-reference reads (raw reads or consensus reads) divided by the total number of reads for that base position. Data from five amplicons covering 417 nucleotides and 12 replicates using the same CP-A DNA source are shown. The average raw read depth was 2.3 × 106 per amplicon and the average consensus read depth was 7700 per amplicon when 30 raw reads with the same barcode was applied as cutoff. (B) Plot showing raw and consensus read error for each base position along with corresponding 95% confidence intervals. (C) Dot plot of total raw and consensus read errors for 13 amplicons and 1042 nucleotides. The average raw read depth was 5.5 × 105 per amplicon and the average consensus read depth was 4700 per amplicon when 10 raw reads with the same barcode was applied as cutoff. The amplicons are ranked from left to right by consensus read depth (Supplementary Figure S4) and the nucleotides within each amplicon are ranked from left to right by their total consensus read error. Consensus reads without any observed errors for a given nucleotide are plotted with half the value of the lowest detected read error. The difference between raw read error and consensus read error at each nucleotide position indicates the relative error correction using SiMSen-Seq. (D) Rare mutation detection in TP53. Pooled plasma DNA from more than 10 individuals and DNA from a clonally derived cell line (CP-A) were analyzed for a single TP53 amplicon using SiMSen-Seq (n = 3–4). The x-axis represents individual nucleotide positions in the amplicon. For each nucleotide position, we identified the most frequent, non-reference (variant) allele and determined both absolute variant count (left side y-axis) normalized to read depth and variant allele frequency (right side y-axis). Error bars indicate 95% confidence intervals for the consensus error observed at each position. Primary tumor DNA with a known TP53 mutation (marked *) was spiked into the plasma DNA at two different allele fractions with 10× separation (blue and red marked bar).
SiMSen-Seq allows rare mutations to be detected in blood plasma samples
To evaluate SiMSen-Seq sensitivity, we spiked primary tumor DNA with known mutations into pooled plasma DNA prepared from >10 individuals without any known disease. For comparison, we also analyzed DNA from the cell line CP-A. Five short amplicons (≤107 base pairs) targeting 252 nucleotides were analyzed (Supplementary Table S1). Figure 4D shows detection of a spiked in TP53 mutation at two different frequencies (0.59 and 0.064%, respectively). In addition to frequency, the absolute number of variants per nucleotide is also indicated in the plot. The upper 95% confidence interval of the control CP-A DNA for that given nucleotide was 0.065%. Two additional spike in mutations are shown in Supplementary Figure S7. In addition to the known spike in mutations we also observed several variants in the plasma DNA at frequencies between 0.10 and 0.64% that did not originate from the primary tumor DNA (Supplementary Figure S7 and Table S2).
DISCUSSION
Incorporating barcodes into NGS libraries using PCR permits background noise reduction and sensitive mutation detection with low DNA inputs. PCR-introduced barcoding applied to a single target sequence was first reported by Kinde et al. using an approach named Safe-SeqS (20). However, Safe-SeqS has not found widespread use and this is probably due to the complex protocol and the fact that it requires a gel-purification step. In our own laboratory, we found that the Safe-SeqS protocol results in large amounts of non-specific PCR product and we were unable to detect specific PCR product on gels when testing five different amplicon designs (Supplementary Table S1). Furthermore, we were reluctant to perform the gel-purification step due to cross-contamination concerns and instead we developed a modified, simpler approach. SiMSen-Seq uses two strategies to reduce non-specific PCR products while simultaneously increasing specific products. The first strategy is the use of a molecular hairpin to protect the barcodes during the initial round of PCR. This prevents the barcodes from participating in mis-priming events, minimizes non-specific PCR products and enables robust formation of the desired product. The second strategy is a standard multiplex pre-amplification approach using low primer concentrations and an elongated PCR extension time in order to compensate. In addition to a greatly simplified protocol without gel purification, SimSen-seq also enables high-level, flexible multiplexing that has not yet been demonstrated with Safe-SeqS. Importantly, as long as the target primer sequence is designed with standard criteria (primer annealing temperature 58–62°C and 20–80% GC content) the hairpin structure is universal for all forward primers. In our experience, failure of any SiMSen-Seq assay could always be traced back to poorly functioning target primers and this can be easily ascertained prior to purchasing primers that incorporate hairpins. When good target primers are selected, we found that all amplicons performed well in SiMSen-Seq, providing reasonably uniform raw read depths and consensus read depths. This was true in the 5-plex, 13-plex and 31-plex data and there is no reason to believe that higher order multiplexing would not perform similarly. However, error reduction by barcoding requires very high sequencing depth and thus can get very expensive depending on the number of targets analyzed. For example, 40 ng total DNA consists of ∼12 000 copies of each sequence. Optimally, 240 000 reads per target are needed to perform error correction using 20 consensus reads. Practically, more reads are needed since not all barcoded molecules for a given target are amplified equally and the uniformity between different amplicons varies. This highlights a major advantage of SiMSen-Seq over both SafeSeqS and, any potential ligation and capture approach, in that it is very flexible and amplicons can be used in combinations with varying levels of multiplexing. Thus, sequencing costs can be minimized by the use of appropriately sized panels designed for specific uses or even for specific samples, such as analysis of plasma DNA in cancer patients where mutations in the tumor are already known. In addition, SiMSen-Seq uses an extremely simple library preparation workflow that is completed within 3 h, eliminating several enzymatic and purification steps that are associated with most NGS protocols, including the Safe-SeqS protocol. Furthermore, the library preparation is highly cost-efficient, since primers and reagents can be purchased individually as needed.
As with any barcoding approach, SiMSen-Seq cannot correct for polymerase-induced errors introduced in the first PCR extension as all daughter molecules will contain the same error and barcode. Furthermore, although SiMSen-Seq does work with two cycles of PCR barcoding (data not shown), we choose to use three cycles as it results in the production of more barcoded template molecules and allows us to inactivate the first PCR with a combined TE buffer dilution and protease digestion step instead of performing a more labor intensive PCR clean-up. Using three cycles does however potentially reduce error correction as polymerase errors in the second PCR extension, initiated by a new barcoded primer, will also be incorporated into all subsequent daughter strands with that barcode. Thus, additional uncorrectable errors (background noise) are introduced using three cycles versus two, but with the benefit of an easier workflow. However, sequencing errors may also be introduced by factors other than the polymerase, including chemically modified nucleotides present in the template DNA and base calling errors that are not dependent on the number of initial PCR cycles (12). Regardless, our experimental setup with SiMSen-Seq was able to clean-up all raw-read hot spot nucleotides, demonstrating that the applied approach is suitable to accurately detect rare sequence variants down to ∼0.1%. This corresponds to 10 molecules or less in most of our analyses.
Supplementary Material
REFERENCES
- 1.ten Bosch J.R., Grody W.W. Keeping up with the next generation: massively parallel sequencing in clinical diagnostics. J. Mol. Diagn. 2008;10:484–492. doi: 10.2353/jmoldx.2008.080027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox E.J., Reid-Bayliss K.S., Emond M.J., Loeb L.A. Accuracy of next generation sequencing platforms. Next Gener. Seq. Appl. 2014;1 doi: 10.4172/jngsa.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo Y.M., Chiu R.W. Genomic analysis of fetal nucleic acids in maternal blood. Annu. Rev. Genomics Hum. Genet. 2012;13:285–306. doi: 10.1146/annurev-genom-090711-163806. [DOI] [PubMed] [Google Scholar]
- 4.Diaz L.A., Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J. Clin. Oncol. 2014;32:579–586. doi: 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.New M.I., Tong Y.K., Yuen T., Jiang P., Pina C., Chan K.C., Khattab A., Liao G.J., Yau M., Kim S.M., et al. Noninvasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin. Endocrinol. Metab. 2014;99:E1022–E1030. doi: 10.1210/jc.2014-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chitty L.S., Lo Y.M. Noninvasive prenatal screening for genetic diseases using massively parallel sequencing of maternal plasma DNA. Cold Spring Harb. Perspect. Med. 2015;5 doi: 10.1101/cshperspect.a023085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsui N.B., Kadir R.A., Chan K.C., Chi C., Mellars G., Tuddenham E.G., Leung T.Y., Lau T.K., Chiu R.W., Lo Y.M. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011;117:3684–3691. doi: 10.1182/blood-2010-10-310789. [DOI] [PubMed] [Google Scholar]
- 8.Murtaza M., Dawson S.J., Tsui D.W., Gale D., Forshew T., Piskorz A.M., Parkinson C., Chin S.F., Kingsbury Z., Wong A.S., et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 9.Tie J., Kinde I., Wang Y., Wong H.L., Roebert J., Christie M., Tacey M., Wong R., Singh M., Karapetis C.S. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015;8:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoque M.O., Lee J., Begum S., Yamashita K., Engles J.M., Schoenberg M., Westra W.H., Sidransky D. High-throughput molecular analysis of urine sediment for the detection of bladder cancer by high-density single-nucleotide polymorphism array. Cancer Res. 2003;63:5723–5726. [PubMed] [Google Scholar]
- 11.Thunnissen F.B. Sputum examination for early detection of lung cancer. J. Clin. Pathol. 2003;56:805–810. doi: 10.1136/jcp.56.11.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diehl F., Schmidt K., Durkee K.H., Moore K.J., Goodman S.N., Shuber A.P., Kinzler K.W., Vogelstein B. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology. 2008;135:489–498. doi: 10.1053/j.gastro.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Milbury C.A., Li C., Makrigiorgos G.M. Two-round coamplification at lower denaturation temperature-PCR (COLD-PCR)-based sanger sequencing identifies a novel spectrum of low-level mutations in lung adenocarcinoma. Hum. Mutat. 2009;30:1583–1590. doi: 10.1002/humu.21112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milbury C.A., Correll M., Quackenbush J., Rubio R., Makrigiorgos G.M. COLD-PCR enrichment of rare cancer mutations prior to targeted amplicon resequencing. Clin. Chem. 2012;58:580–589. doi: 10.1373/clinchem.2011.176198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogelstein B., Kinzler K.W. Digital PCR. Proc. Natl. Acad. Sci. U.S.A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrett A.N., McDonnell T.C., Chan K.C., Chitty L.S. Digital PCR analysis of maternal plasma for noninvasive detection of sickle cell anemia. Clin. Chem. 2012;58:1026–1032. doi: 10.1373/clinchem.2011.178939. [DOI] [PubMed] [Google Scholar]
- 17.Taly V., Pekin D., Benhaim L., Kotsopoulos S.K., Le Corre D., Li X., Atochin I., Link D.R., Griffiths A.D., Pallier K., et al. Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin. Chem. 2013;59:1722–1731. doi: 10.1373/clinchem.2013.206359. [DOI] [PubMed] [Google Scholar]
- 18.Forshew T., Murtaza M., Parkinson C., Gale D., Tsui D.W., Kaper F., Dawson S.J., Piskorz A.M., Jimenez-Linan M., Bentley D., et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3003726. 136ra168. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012;109:14508–14513. doi: 10.1073/pnas.1208715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinde I., Wu J., Papadopoulos N., Kinzler K.W., Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2011;108:9530–9535. doi: 10.1073/pnas.1105422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q., Vijaya Satya R., Lewis M., Randad P., Wang Y. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics. 2015;16:589. doi: 10.1186/s12864-015-1806-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Narayan A., Carriero N.J., Gettinger S.N., Kluytenaar J., Kozak K.R., Yock T.I., Muscato N.E., Ugarelli P., Decker R.H., Patel A.A. Ultrasensitive measurement of hotspot mutations in tumor DNA in blood using error-suppressed multiplexed deep sequencing. Cancer Res. 2012;72:3492–3498. doi: 10.1158/0008-5472.CAN-11-4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bettegowda C., Sausen M., Leary R.J., Kinde I., Wang Y., Agrawal N., Bartlett B.R., Wang H., Luber B., Alani R.M., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6 doi: 10.1126/scitranslmed.3007094. 224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson D., Akrap N., Svec D., Godfrey T.E., Kubista M., Landberg G., Stahlberg A. Properties of targeted preamplification in DNA and cDNA quantification. Expert Rev. Mol. Diagn. 2015;15:1085–1100. doi: 10.1586/14737159.2015.1057124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brownie J., Shawcross S., Theaker J., Whitcombe D., Ferrie R., Newton C., Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.