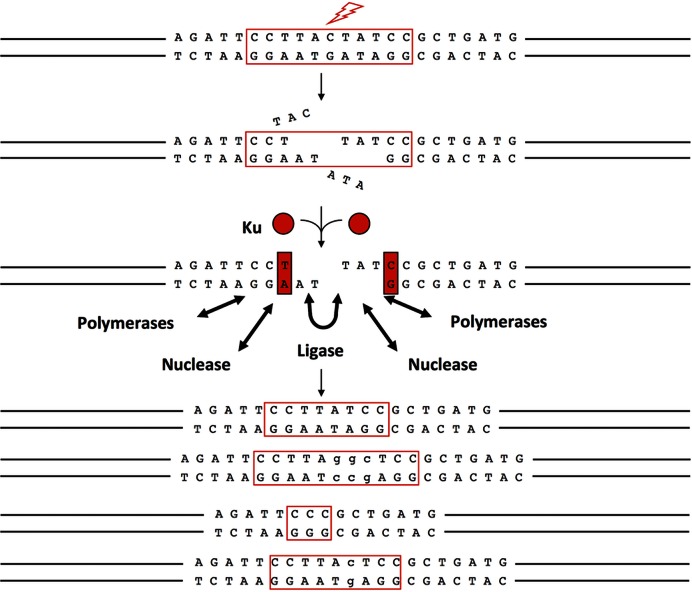
Figure 4.
Artemis is involved in DNA end repair via the NHEJ pathway. Natural causes of pathologic double-strand breaks (DSBs) are expected to generate heterogeneous, incompatible DNA ends with little or no terminal microhomology. Ku (red circle and red rectangle) is the most abundant DNA end binding protein in eukaryotic cells and can slide onto DNA ends that have diverse configurations (38). Once Ku is bound to the DNA end, it can improve the binding equilibrium of the nuclease, polymerases and ligase of NHEJ. The nuclease, polymerases and ligase appear capable of binding to and functioning at a DNA end without Ku, but the binding to the DNA end is tighter when Ku is present. The most clearly identified nuclease thus far is the Artemis:DNA-PKcs complex. The polymerases for NHEJ include the POL X polymerases, pol μ and pol λ (39). The ligase complex of NHEJ consists of XLF, PAXX, XRCC4 and DNA ligase IV (40–42). The bottom portion of the diagram shows four equally plausible outcomes for the joining (the junction is highlighted in a red box). There are hundreds of other possible joining outcomes even for one pair of starting DNA ends with the same end configuration as that shown. This heterogeneity in outcome is in addition to the heterogeneity in the end configuration generated by the original breakage process at that very same set of phosphodiester bonds within the DNA duplex. Thus, there is heterogeneity in the generation of the broken DNA ends and heterogeneity in how these ends are repaired.