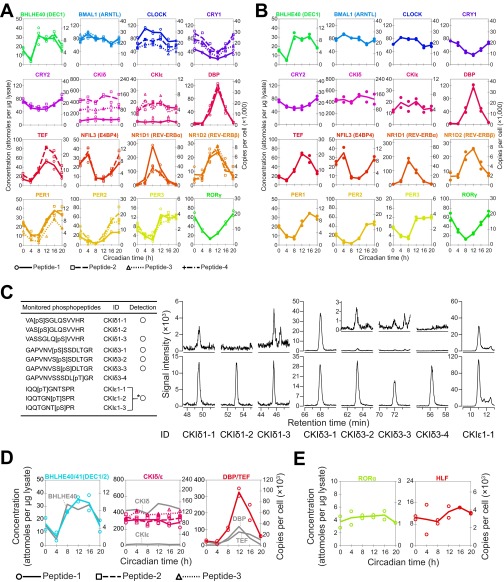
Fig. S5.
A summary of the expression levels of each peptide. (A, B, D, and E) Concentrations are expressed as attomol per microgram of lysate (Left axis) or copies per cell (Right axis). Copy numbers were calculated by estimating a total amount of proteins per cell is 0.5 ng (19). (A) Quantification of each of the peptides from a protein. Time courses of the quantified peptides arranged according to their parental proteins are shown. The correspondence of detected peptides to plots is shown in Dataset S2. (B) Time course of the quantified proteins calculated from two biological replicates. Closed circles indicate average peptide concentrations derived from same protein in each biological replicate. (C) SRM detection of phosphorylated peptide-1 and peptide-3 from CKIδ and peptide-3 from CKIε. Monitored phosphopeptides are summarized in the table (Left). SRM chromatograms (Right) corresponding to peptides derived from mouse liver (Upper) and isotope-labeled internal standards (Lower) are shown. An asterisk (*) indicates that the peptides were not separated by chromatography and therefore could not be differentiated from each other in the chromatogram. (D) Quantification of peptides for common region of two circadian clock proteins. Time courses of the quantified peptides for common region of DEC1 (BHLHE40) and DEC2 (BHLHE41), CKIδ and CKIε, and DBP and TEF are shown. The correspondence of detected peptides to plots is shown in Dataset S2. Gray lines indicate the time course of quantified parental proteins. (E) Quantification of peptides with low signal intensities. Time courses of the peptides for HLF and RORα are shown.