Fig. 1.
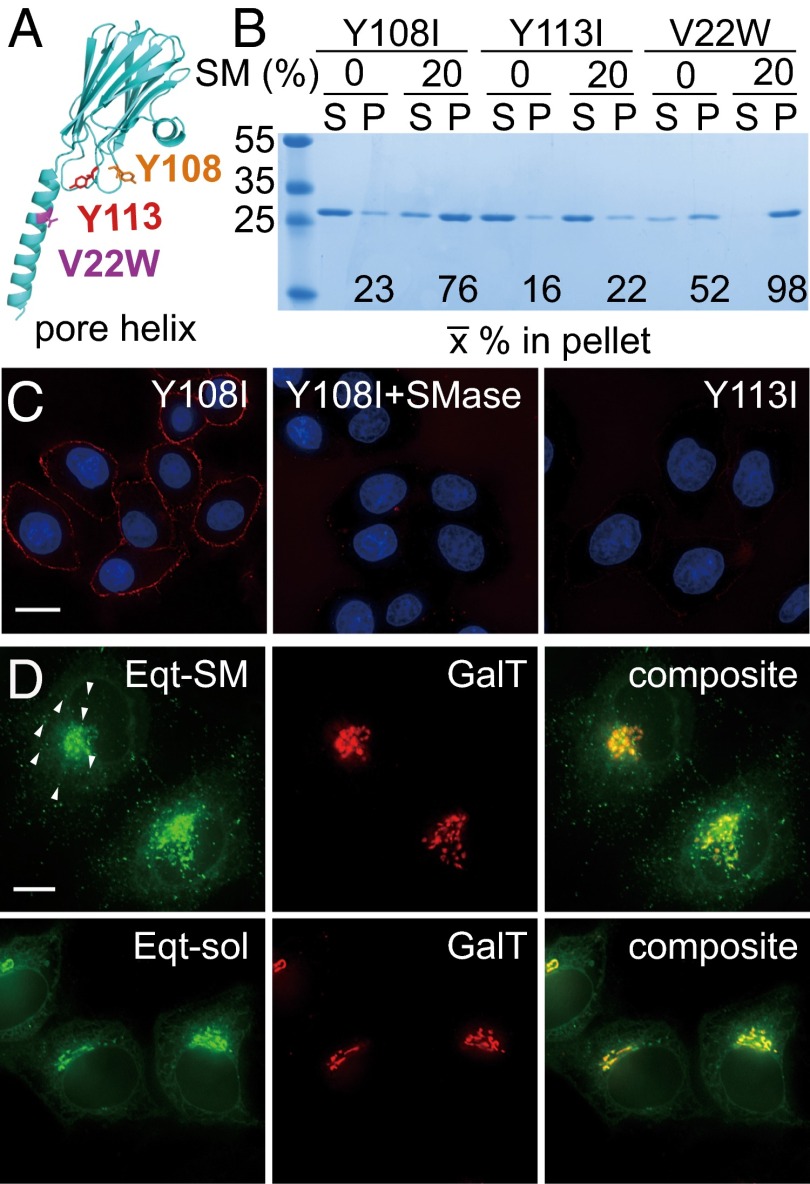
Membrane-binding properties and localization of Eqt derivatives. (A) Location of mutations introduced into Eqt. The structure of an actinoporin (FraC) protomer in its oligomeric pore-forming conformation (28) is rendered, with mutations that we introduced indicated. A residue on the N-terminal helix, V22, which was changed to tryptophan to ablate pore formation, is shown in purple. Y108, which was changed to isoleucine in Eqt-SM, is in orange, and Y113, which was changed to isoleucine in Eqt-sol, is in red. (B) Vesicle-binding assays. The indicated Eqt proteins were incubated with vesicles containing 20% SM or phosphatidylcholine (and 20% cholesterol), and the vesicles were collected by centrifugation. Bound pellet (P) and unbound supernatant (S) fractions were visualized by Coomassie blue staining and quantified. The mean values of three independent experiments are shown. Molecular mass standards (kDa) are indicated on the left. The Y108 and Y113 mutants also include the V22W mutation. (C) Eqt-V22W,Y108I recognizes SM in the plasma membrane of intact cells. Recombinant FLAG epitope-tagged Eqt-Y108I or Eqt-Y113I was incubated with HeLa cells that had been incubated with SMase or mock-treated. Cells were then washed, fixed, and incubated with anti-FLAG and labeled secondary antibodies. (D) Localization of Eqt-SM and Eqt-sol in HeLa cells. Plasmids encoding the indicated proteins were transfected into HeLa cells and visualized by deconvolution florescence microscopy at 16 h after transfection. The arrows point to examples of cytoplasmic puncta that contain Eqt-SM. Maximum projections of z series are shown. (Scale bar: 10 μm.)