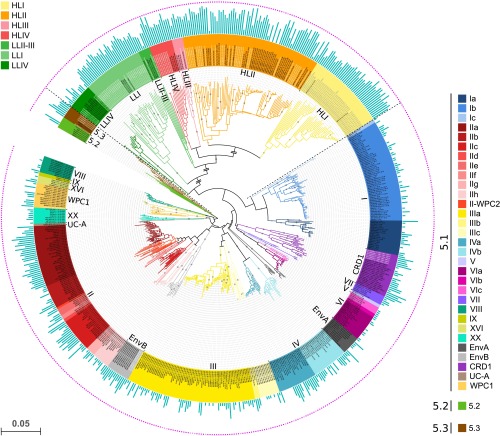
Fig. 1.
Neighbor joining tree of Synechococcus and Prochlorococcus lineages based on petB gene sequences from both isolates and environmental sequences. Diamonds at nodes indicate bootstrap support over 70%. Taxonomic assignments are given by the color codes at clade level for Prochlorococcus (top left corner) and clade (e.g., V, CRD1) or subclade (e.g., Ia-c) for Synechococcus (right side). Sequences were named after ID_subcluster_clade_subclade_ESTU for Synechococcus ID_LL or HL_clade_ESTU for Prochlorococcus. The outer pink ring indicates that the corresponding sequence in the tree was the best hit of at least one Tara Oceans picocyanobacterial read, and the inner blue bar plot shows the log2 of the number of metagenomic reads recruited for this sequence (range: 0–10.84). Sequences in black letters correspond to the initial reference database and those in white or light gray letters to newly assembled petB sequences from Tara Oceans metagenome reads. The scale bar represents the number of substitutions per nucleotide position. For improved readability, the length of three Prochlorococcus branches was reduced, as indicated by double slashes. Prochlorococcus clade assignment is as in ref. 66, whereas for Synechococcus subcluster 5.1, subclade assignments are as in ref. 67 for WPC1 and WPC2 and as in ref. 12 for all other clades.