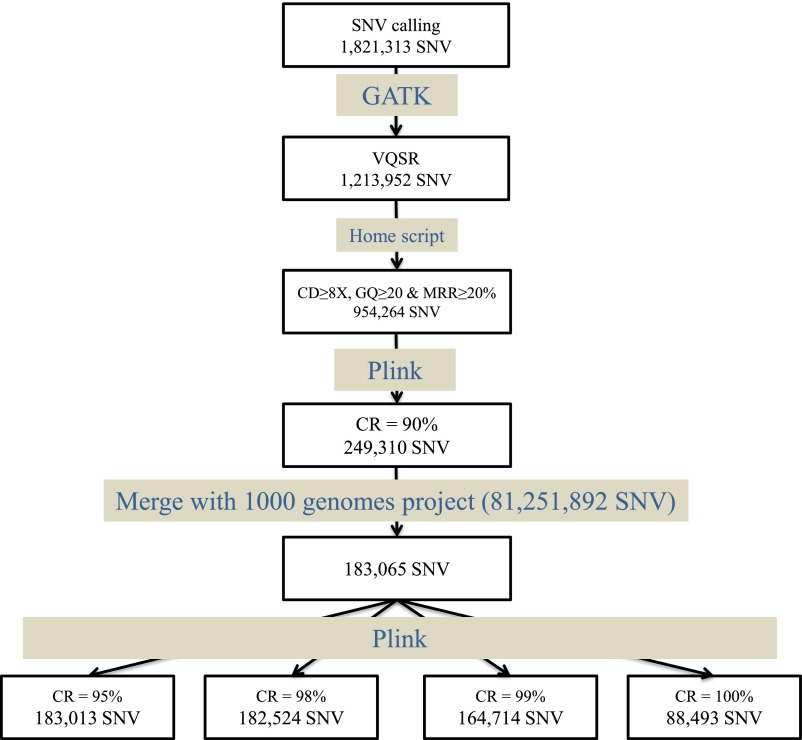
Fig. S1.
Flowchart summarizing the entire WES quality process. First, genotypes for all positions with an SNV (heterozygous or homozygous for the alternative allele) for at least one individual were called. Then, only variants passing the GATK Variant Quality Score Recalibrator (VQSR) filters were kept. Genotypes with a low coverage, genotype quality, or minor read ratio (for heterozygous) were filtered out. Finally, variants with a CR < 90% over our sample of 110 individuals were removed. After merging with the 1000 Genomes Project data, in total, 183,065 SNVs were retained for our analyses, for which we considered four levels of CR between 95% and 100%.