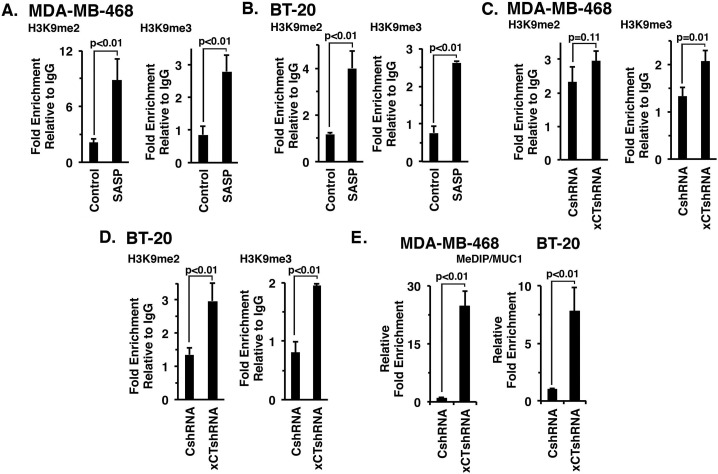
Figure 6. Targeting xCT induces histone and DNA methylation of the MUC1 promoter.
A. and B. MDA-MB-468 (A) or BT-20 (B) cells were treated with 1.0 mM or 1.5 mM SASP for 72 h, respectively. Soluble chromatin from control and SASP treated cells was precipitated with anti-H3K9me2, anti-H3K9me3 or a control IgG. The final DNA samples were amplified by qPCR with pairs of primers for the MUC1 promoter region or control region from the GAPDH promoter. The results (mean±SD of 3 determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (untreated cells, assigned a value of 1). C. and D. Soluble chromatin from MDA-MB-468/CshRNA, MDA-MB-468/xCTshRNA (C), BT-20/CshRNA and BT-20/xCTshRNA (D) cells was precipitated with anti-H3K9me2, anti-H3K9me3 or a control IgG. The final DNA samples were amplified by qPCR with pairs of primers for the MUC1 promoter region or control GAPDH promoter. The results (mean±SD of 3 determinations) are expressed as the relative fold enrichment compared with that obtained with the IgG control (CshRNA cells, assigned a value of 1). E. Genomic DNA from the indicated MDA-MB-468 (left) and BT-20 (right) cells was subjected to immunoprecipitation of methylated DNA (MeDIP) and the precipitates were analyzed by qPCR of the MUC1 gene promoter. The results (mean±SD of 3 determinations) are expressed as relative fold enrichment compared to that obtained from CshRNA expressing cells (assigned a value of 1).