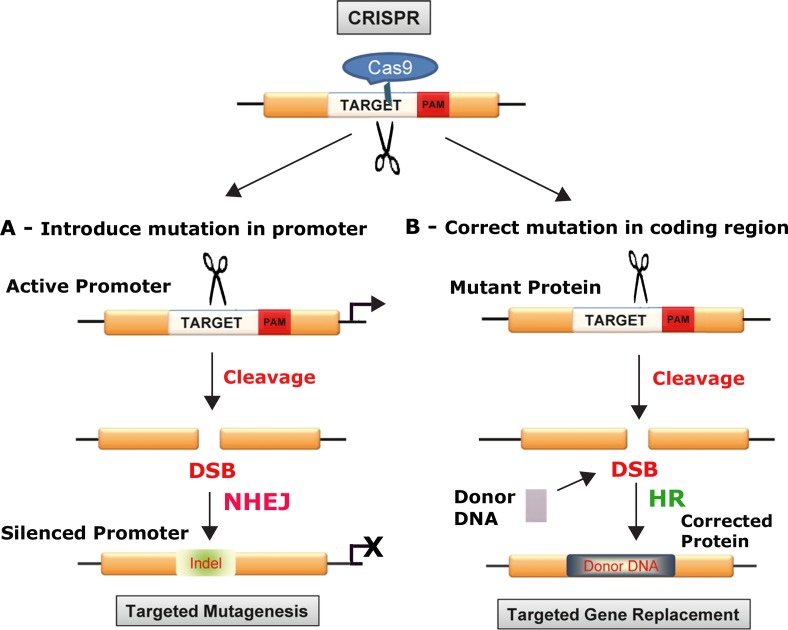
Figure 1. Schematic of gene disruption (A) and correction (B) approaches with CRISPR/Cas9: In the top panel, the relationship of the target DNA sequence, the Protospacer Adjacent Motif (PAM) trinucleotide sequence and Cas9 protein to the scission of the target.
Cas9 cuts both strands of DNA causing a DSB, which lies 3-4 nucleotides upstream of the PAM sequence, which can be used to either disrupt DNA by targeted mutagenesis (A) or replace and correct a mutated gene as shown below (B). CRISPR/Cas9 can be used to silence a promoter. A. double-stranded DNA break is introduced by specific cleavage and this is repaired by the error-prone process of nonhomologous end-joining DNA repair (NHEJ), which introduces InDel mutations that can disrupt the function of a promoter region. This method can be used to disrupt oncogenes, e.g., myc that is expressed at a high level due to translocation into immunoglobulin loci in Burkitt's lymphomas. B. CRISPR/Cas9 can be used to repair and correct a mutated gene. Double-strand DNA breaks are introduced by specific cleavage but in this case repair is mediated by the high-fidelity mechanism of homologous recombination-directed DNA repair (HR). This can be used to repair mutated tumor suppressor genes and restore the wild-type sequence and function.