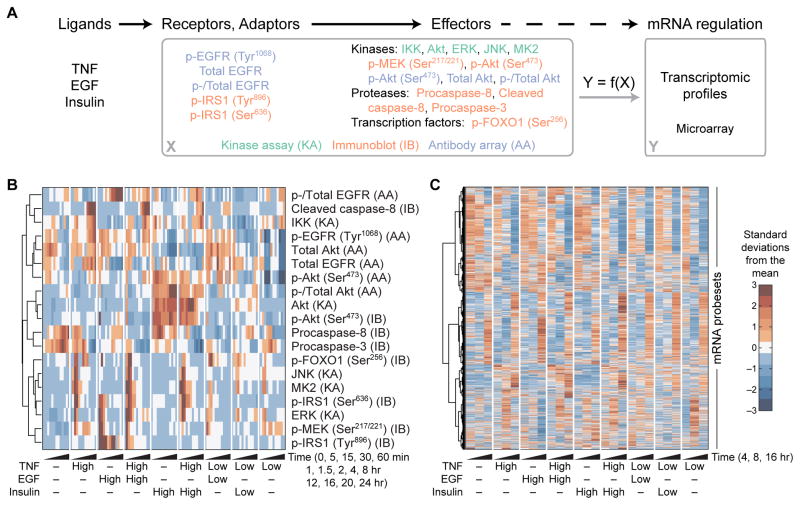
Fig. 1. A compendium of ligand--induced signals and transcriptional responses.
(A) Overview of the experimental design. HT-29 cells were pretreated with IFNγ, stimulated with various combinations and concentrations of TNF, EGF, and insulin, and profiled for the indicated signaling receptors, adaptors, and effectors by kinase assay (KA), immunoblot (IB), or antibody array (AA) and for the associated transcriptomic signatures by microarray. The goal is to determine whether global ligand-induced mRNA regulatory states (Y) can be predicted from the upstream signaling network activation (X). (B) Hierarchical clustering of the signaling compendium for saturating (High) and subsaturating (Low) concentrations of TNF, EGF, and insulin (42, 43). Data are means of n = 3 to 6 independent biological replicates. (C) Hierarchical clustering of the dynamic transcriptomic responses resulting from the ligand combinations in (B). Data are means of n = 2 independent biological replicates.