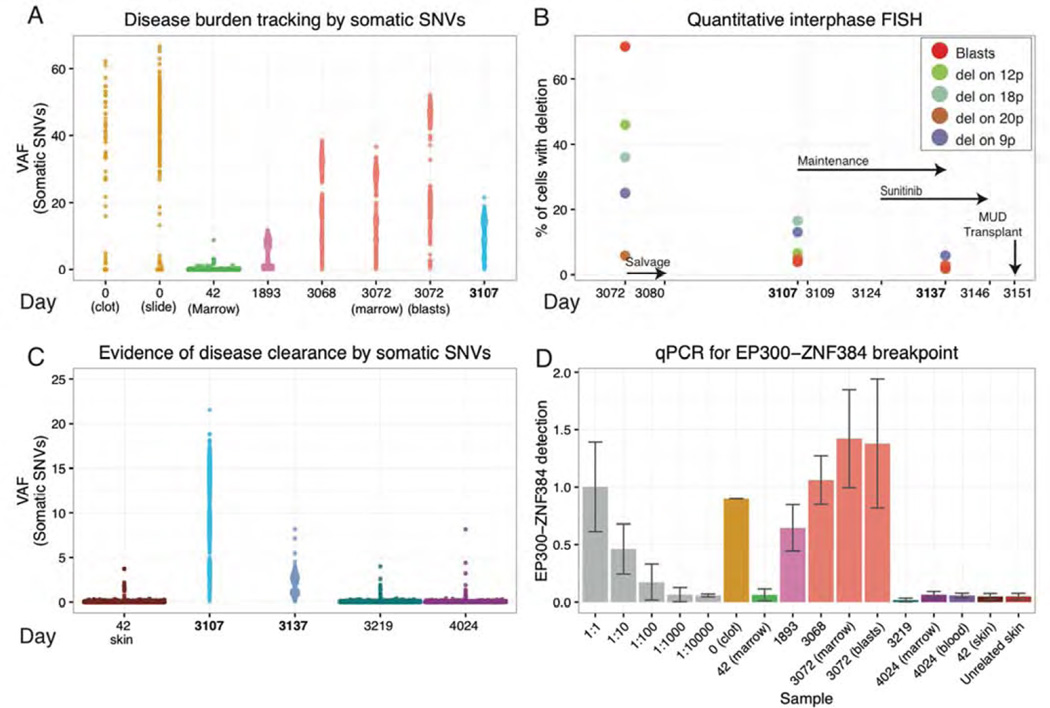
Figure 4. Personalized genomic assays for assessment of tumor burden and response to treatment.
Disease burden and evidence for disease clearance prior to the final MUD allograft were assessed by three orthogonal approaches: VAFs from deep custom capture sequencing for validated somatic SNVs, quantitative FISH for four somatic large-scale deletions, and qPCR of a somatic EP300-ZNF384 translocation breakpoint found to be a possible early/initiating event in this tumor. Time points assayed range from the day of initial diagnosis (day 0) to the patient’s most recent follow-up biopsy obtained ~2.5 years into his current sustained remission (day 4,024). (A) VAFs of 1,588 somatic variants are displayed for eight samples across six time points from diagnosis of the primary B-ALL (day 0), first relapse (day 1,893), and second relapse (day 3,068/3,072). A 40× coverage filter was applied to the primary samples further limiting variants to approximately 581 and 270 for the slide and clot, respectively (Methods). (B) Results from quantitative FISH (Methods) are displayed for four structural deletions utilized to assess tumor burden at the second relapse and following weeks. Refer to Table S3 for details on each deletion assayed. Key treatment time points are indicated with black arrows. (C) VAFs of somatic variants used to assess disease clearance at day 3,219 and day 4,024 are contrasted with their VAFs during refractory disease post salvage therapy (day 3,107) and in the normal skin sample obtained during the first remission (day 42). The sample from day 3,137 was obtained 13 days after initiation of sunitinib, and demonstrates a highly significant reduction in average VAFs compared to that of day 3,107 (p-value < 2.2e-16; n=1,588; Wilcox signed-rank test). (D) qPCR data representing abundance of the EP300-ZNF384 breakpoint in genomic DNA is displayed for a dilution series of the day 3,072 relapse sample with an unrelated skin sample (grey), six disease samples designated by the day of collection, three samples obtained after the MUD transplant, a matched skin normal obtained in first remission (day 42), and a skin sample from an unrelated donor who did not have this translocation (used as a reference to measure assay “noise”) Note the non-linear, square root transformed values used for display purposes (Methods).