Figure 6.
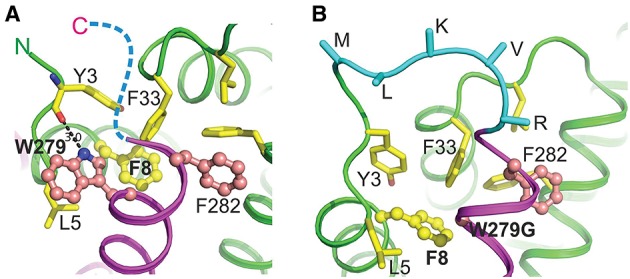
Predicted molecular interactions between the C-terminus of YopN and the N-terminus of TyeA in the YopN-TyeA complex and YopN-TyeA fusion protein. Cartoon diagram of a fragment of the crystal structure of the YopN-TyeA complex (RCSB PDB accession code 1XL3; A). The C-terminal helix of YopN is painted in magenta and TyeA is shown in green. Two C-terminal residues of YopN, significantly contributing to the binding interface, Trp279 and Phe282, are shown as balls-on-sticks with carbon and nitrogen atoms painted in pink and blue, respectively. Each of these two residues contributes about 10% to the total interactive area (1099 Å2), establishing hydrophobic interactions with TyeA. In addition, the nitrogen atom of the side chain of Trp279 forms a hydrogen bond with the main chain carbonyl group of Tyr3 in TyeA. Residues that interact with Trp279 and Phe282 are shown in sticks or balls-on-sticks (Phe8) with carbon, nitrogen, and oxygen atoms painted in yellow, blue, and red, respectively. The hydrogen bond between Trp279 and Tyr3 is shown with a dashed line (length 3.0 Å). Our study demonstrates the pivotal role of Trp279 of YopN and Phe8 of TyeA in the YopN-TyeA binding. The ten-residue C-terminus of YopN is unstructured (indicated by a blue dashed line) and, as we show here, plays no role in the binding. Cartoon diagram of a model of the YopN-TyeA fusion protein as a consequence of a mutated yopN allele containing an engineered in cis +1 frameshift mutation immediately downstream of codon 278 (Amer et al., 2013; B). The model was produced based on the crystal structure of the YopN-TyeA complex using program O. The connecting loop (cyan) was designed based on the search of loop library, keeping high restrains for stereochemistry. The side chains of residues at the C-terminus that are altered due to the +1 frame-shift were modeled using the most frequently found rotamer conformations. Only Cα and Cβ atoms are shown for the connecting loop residues. The interactive residues are shown as in (A). The figure was generated by PYMOL (http://www.pymol.org/).
