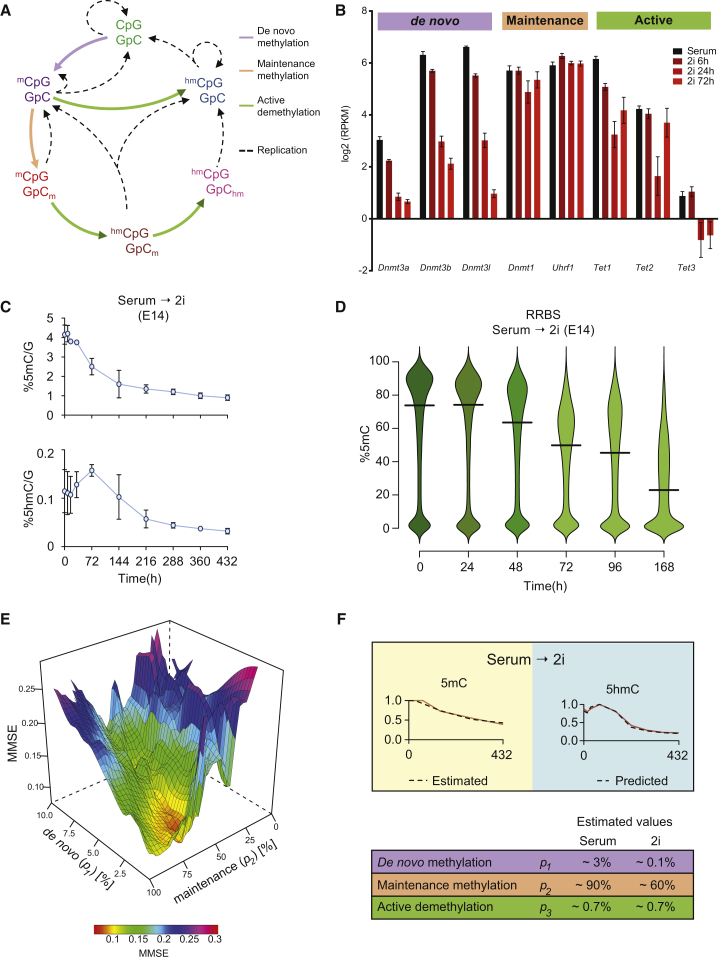
Figure 1.
Dynamic Regulation of 5mC and 5hmC during Serum-to-2i Conversion of Mouse ESCs
(A) Schematic representation of cytosine methylation/demethylation cycle. Different forms of modified CpG dyads and the corresponding processes are indicated.
(B) Expression levels of genes involved in the DNA methylation machinery during serum-to-2i transition. Error bars indicate mean ± SD from three biological replicates.
(C) Percentage of 5mC (top) and 5hmC (bottom) as measured by LC-MS in E14 during serum-to-2i conversion. Error bars indicate mean ± SD from three biological replicates.
(D) Percentage of 5mC methylation as measured by RRBS in E14 during serum-to-2i conversion. Horizontal bars represent the median values.
(E) Graphical representation of the mathematical modeling. The minimum mean square error (MMSE) is plotted against different values for de novo methylation (p1) and maintenance methylation (p2).
(F) Overlay of the mathematical model predictions (dotted line) with real measurements (red line) obtained from LC-MS in E14 during serum-to-2i conversion. The table summarizes the results of the mathematical modeling, showing the values estimated for p1, p2, and p3 in the serum steady state and in 2i. p1, proportion of unmethylated CpGs that become hemi-methylated per average cell division; p2, proportion of hemi-methylated CpGs that become fully methylated per average cell division; and p3, proportion of methylated CpGs that become hydroxymethylated per average cell division.