Figure 2.
Global Identification of Loci Repositioning in C2C12 Myogenesis
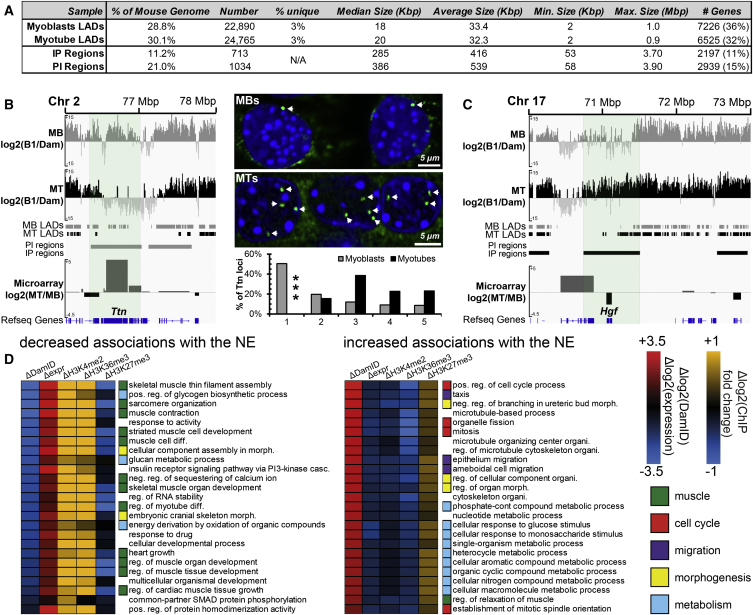
(A) Table summarizing parameters of identified LADs and regions showing increased (IP) and decreased (PI) association with the nuclear periphery.
(B) Example genome browser view of the Ttn locus with representative images and direct quantification of the position of Ttn loci in 100 nuclei (green) by FISH in MBs and MTs. ∗∗∗p < 0.001 comparing locus position in MTs to MBs using χ2 test.
(C) Genome browser view for the genomic region surrounding Hgf showing DamID signal intensities, identified LADs, IP and PI regions, and microarray gene expression changes for MBs and MTs.
(D) Heatmap displaying average Δlog2 DamID, Δlog2 expression, and Δlog(fold change) for indicated histone modifications values between MTs and MBs for genes within GO term categories significantly enriched in PI activated genes and IP repressed genes. Myogenic alterations to histone modifications associated with transcriptionally active genes (H3K4me2 and H3K36me3) and the transcriptional repression-associated H3K27me3 were extracted from Asp et al. (2011). The Δlog2(DamID) value was calculated by subtracting the average log2(Lamin B1/Dam) value in a 100 kb window surrounding the gene in the MB sample from the MT sample. The ChIP-seq values were determined for each histone modification by subtracting the average signal across the gene body in the MB sample from the MT sample.
See also Figures S2 and S3.