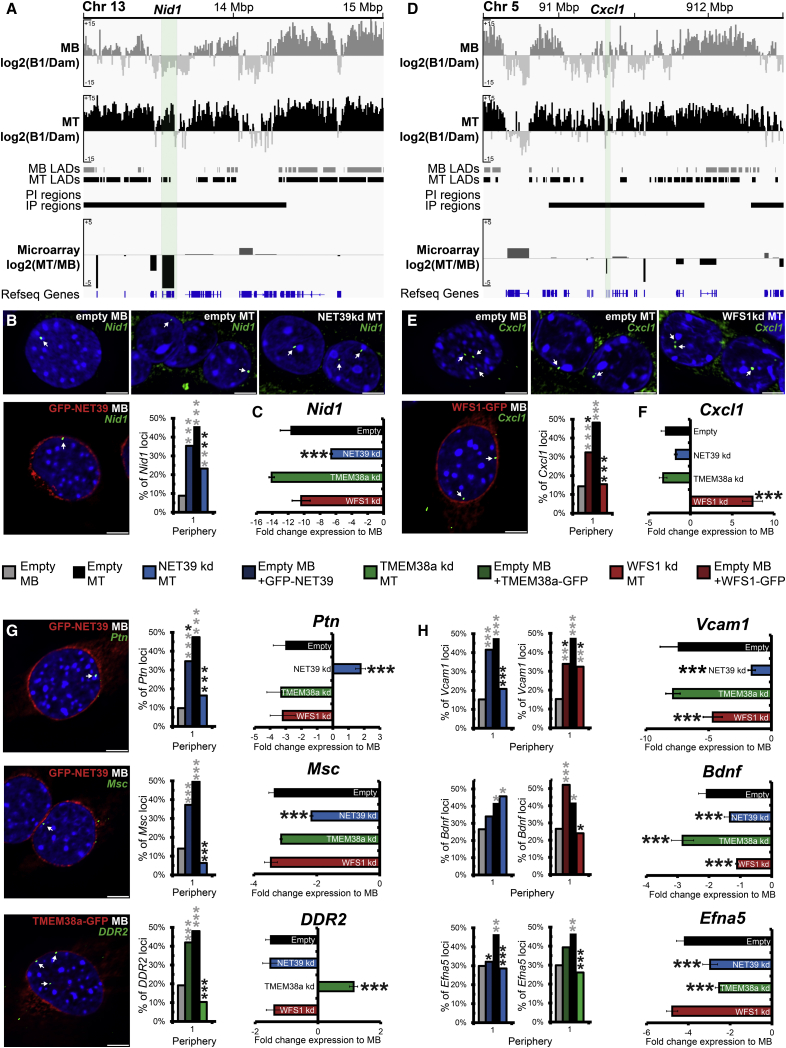
Figure 5.
FISH and Microarray Analysis in NET KDs Confirms NET-Directed Repositioning and Expression Regulation
(A and D) Genome browser views of myogenic DamID and gene expression changes of the Nid1 (A) and Cxcl1 loci (D).
(B and E) FISH analysis of Nid1 (B) and Cxcl1 (E) loci position in control MBs and MTs, NET-depleted MTs, and NET-overexpressing MBs.
(C and F) Histogram of Nid1 (C) and Cxcl1 (F) gene expression changes relative to MBs in control and NET-depleted MTs.
(G and H) Similar microarray and FISH analysis of genes affected uniquely by depletion of a NET (Ptn, Msc, and DDR2) (G) and by depletion of multiple NETs (Vcam1, Bdnf, and Efna5) (H). For microarray data, error bars represent SD over three biological repeats, while statistics represent false discovery rates (FDR) between sample and an empty vector-treated control. For FISH, loci position was determined in 50–100 nuclei for each sample. For quantification statistics, the position of loci in the indicated sample was compared to the empty-vector MBs (gray asterisks) or MTs (black asterisks) using χ2 tests. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
All FISH statistics are in Figure S6. The scale bars represent 5 μm.