Figure 3.
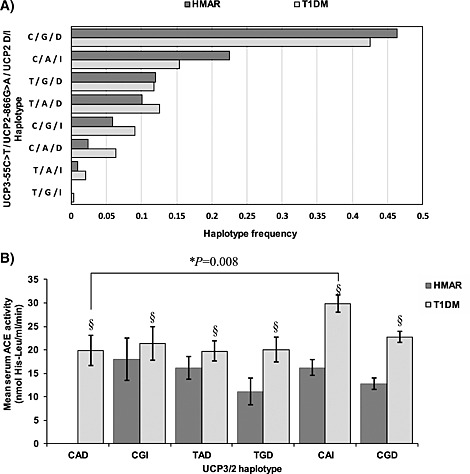
A: The UCP3‐55C>T, UCP2‐866G>A and UCP2D/I haplotype frequencies are shown in descending order of frequency for both the HMAR and T1DM studies. The most common haplotype is the UCP3‐55C/UCP2‐866G/UCP2D (CGD) allele. Subsequent analysis was confined to common haplotypes with a frequency greater than 0.05. B: Predicted UCP3‐55C>T, UCP2‐866G>A and UCP2D/I haplotype effects on age‐adjusted serum ACE (sACE) activity in the HMAR and T1DM study subjects. Data are mean ± SD for each single allele effect. Haplotypes with a frequency less than 0.05 are not represented in the figure because of the very small group size and resulting wide standard deviation for sACE activity. §There was a significant haplotype effect in predicting sACE activity overall in the T1DM subjects (P = 0·01), but not in the HMAR study. *The bracket shows the comparison in the T1DM study between those individuals with the CAD haplotype who had the lowest mean sACE activity and those with the CAI haplotype who had the highest mean sACE activity. A SNP change from the UCP3/2 CAD to CAI haplotype led to a significant mean increase in sACE activity of 10·0 nmol his‐leu/ml/min in the T1DM study (P = 0·008). Individuals with the CAD haplotype in the HMAR study also had the lowest sACE activity but are not shown owing to the low sample size.
