Figure 3.
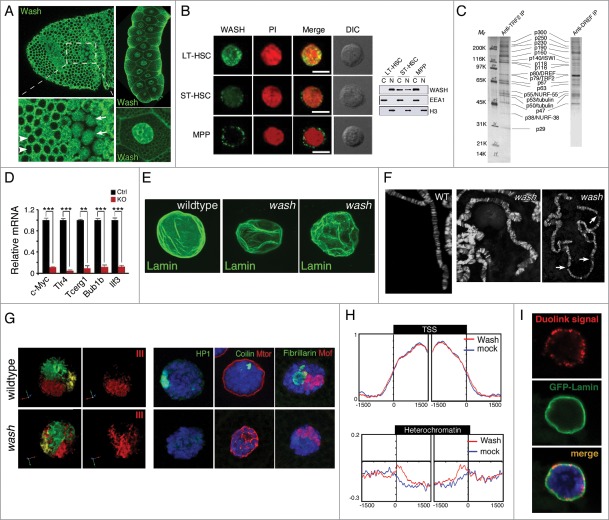
Nuclear localized WASH family proteins function as regulators of nuclear morphology and as transcription factors. (A) Wash accumulates in the nucleus of Drosophila cells in a temporally and spatially specific manner. Confocal projection of stage 7 embryos immunostained with anti-Wash show nuclear localization in specific mitotic domains (arrows) while remaining cytoplasmic in others (arrowheads) (left). Confocal projections of the nuclei of the salivary glands of 3rd instar larvae immunostained with anti-Wash shows nuclear enrichment (right). (B) WASH exhibits different sub-cellular localizations in different sub-populations of haematopoietic stem cells. Micrographs (left) and western blot analysis of lysates (right) of WASH expression in long-term haematopoietic stem cells (LT-HSC; nuclear), short-term haematopoietic stem cells (ST-HSC; nuclear and cytoplasmic) and multipotent progenitor cells (MPP; cytoplasmic). Micrographs are co-stained with PI for nuclear visualization and DIC views are shown (left). (C) Purification of the Drosophila TRF2 complex showing the presence of Wash and its SHRC. Co-immunoprecipitation by TRF2 or DREF monoclonal antibodies purifies a complex containing Wash (p63), SWIP (p116) and Strumpellin (p118), in addition to ISWI, DREF, TRF2, and tubulin. (D) WASH knockout in LT-HSCs leads to reduced c-Myc expression, as well as reduced expression of its transcriptional targets. Bar plot graphs of gene expression levels measured by qPCR showing significantly decreased expression of c-Myc, and its target genes Tlr4, Tcerg1, Bub1b, and Ilf3, in cell-sorted WASH KO LT-HSCs. (E–G) wash mutant salivary glands have altered nuclear morphology compared to wildtype. Confocal projections of wildtype vs. wash mutant nuclei immunostained with anti-Lamin antibody shows crinkled, non-spherical wash nuclei (E). Micrographs of wildtype versus wash mutant salivary gland polytene chromosomes show misalignment and improper banding, as well as extremely fragile chromosomes (F). Three-Dimensional reconstruction of wildtype vs. wash mutant salivary gland nuclei hybridized with chromosome-specific paints (X chromosome: yellow, 2nd chromosome: green, 3rd chromosome: red) showing less compact chromosome territories in wash mutant nuclei (G, left). Micrographs of wildtype verses wash mutant salivary gland nuclei immunostained with anti-HP1 (green; heterochromatin; left), anti-Coilin (green; Cajal bodies; middle), anti-Mtor (red; nuclear envelope protein; middle), anti-Fibrillarin (green; nucleolus; right), anti-MOF (red; X-chromosome; right) showing disrupted nuclear sub-compartments in wash mutant nuclei (G, right). (H) Wash increases chromatin accessibility in heterochromatin regions. Distribution of M.SssI-based chromatin accessibility in control RNAi and wash RNAi treated Drosophila S2 cells, showing increased accessibility in wash knockdown with no affect on transcription start site (TSS) chromatin regions. (I) Wash interacts with B-type Lamin at the nuclear periphery. Duolink proximity ligation assay in Drosophila salivary glands expressing GFP-Lamin using anti-Wash and anti-GFP antibodies shows amplification of Duolink signal at the nuclear periphery. Amplification signal is only observed when the 2 antibodies examined are within 30 nm. Permissions. (A, right) and (E–I) Reprinted from Current Biology, 25(6), Verboon et al., Wash Interacts with Lamin and Affects Global Nuclear Organization, pp. 804-810.54 © Elsevier. Reproduced by permission of Elsevier. Permission to reuse must be obtained from the rightsholder. (B, D) Reprinted with permission from: © Rockefeller University Press. Reproduced by permission of Rockefeller University Press. Permission to reuse must be obtained from the rightsholder. Reprinted from Xia et al. Journal of Experimental Medicine. 211:2119-2134. doi:10.1084/jem.20140169.53 (C) © Macmillan Publishers Ltd: Nature. Reproduced by permission of Macmillan Publishers Ltd: Nature. Permission to reuse must be obtained from the rightsholder. Hochheimer et al., Nature 420:439–45.58