Figure 6. Model comparing the state transitions on coding and non‐coding RNA transcripts.
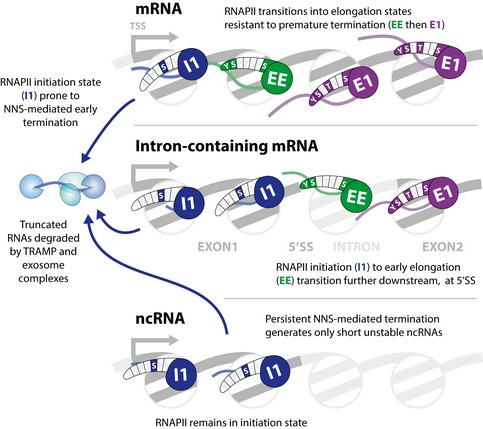
Yeast genes typically have a well‐ordered nucleosome close to the transcription start site. On both mRNA and ncRNA genes, RNAPII generally in an initiation state (I1 and I2; shown as I1) while traversing the first nucleosome. This state favors recruitment of nuclear RNA surveillance machinery, including the NNS termination complex and the TRAMP–exosome degradation system. On mRNAs, RNAPII then transitions into the early elongation state (EE) followed by the major elongation states (E1 to E3; shown as E1). On intron‐containing mRNAs, the transition from initiation of early elongation states is displaced further 3′. On ncRNAs, the initiation state persists and the failure to transition to an elongation states (EE and E1) favors termination and transcript degradation.
