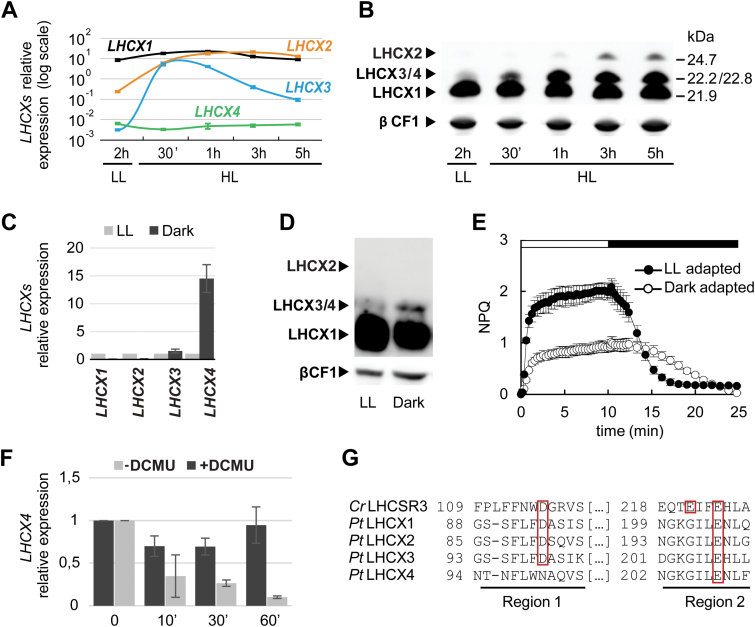
Fig. 1.
Light and dark regulation of P. tricornutum LHCXs. Analysis of the four LHCX transcripts by qRT-PCR (A) and of LHCX proteins (B) by western blotting in cells adapted to low light (LL) (12L/12D cycles), after exposure to LL for 2h then to high light (HL) for 30min, 1h, 3h, or 5h. mRNA levels were quantified by using RPS as the reference gene (A). Proteins were detected using the anti-LHCSR antibody which recognizes all the PtLHCXs (arrowheads) and the anti-βCF1 antibody as loading control (B). Cells adapted to darkness for 60h were compared with those grown in LL for the analysis of LHCX transcripts (C), proteins (D), and NPQ (E). Relative transcript levels were determined using RPS as a reference, and values were normalized to gene expression levels in LL. LHCX proteins were detected as in (B). The horizontal bar in (E) indicates when the actinic light was on (white) or off (black). (F) LHCX4 mRNAs in 60h dark-adapted cells (Time 0) and in response to 10min, 30min, or 1h of blue light (1 µmol m−2 s−1), in the presence (black) or absence (grey) of the inhibitor DCMU. Transcript levels were quantified by using RPS as the reference, and normalized to gene expression levels in the dark. Error bars represent ±SD of three technical replicates from one representative experiment in (A), and ±SD of three biological replicates in (C), (E), and (F). (G) Alignment of regions 1 and 2 of the Chlamydomonas reinhardtii LHCSR3 and P. tricornutum LHCX1, 2, 3, and 4 protein sequences. The boxes indicate the pH-sensing residues conserved between the LHCXs and LHCSR3. (This figure is available in colour at JXB online).