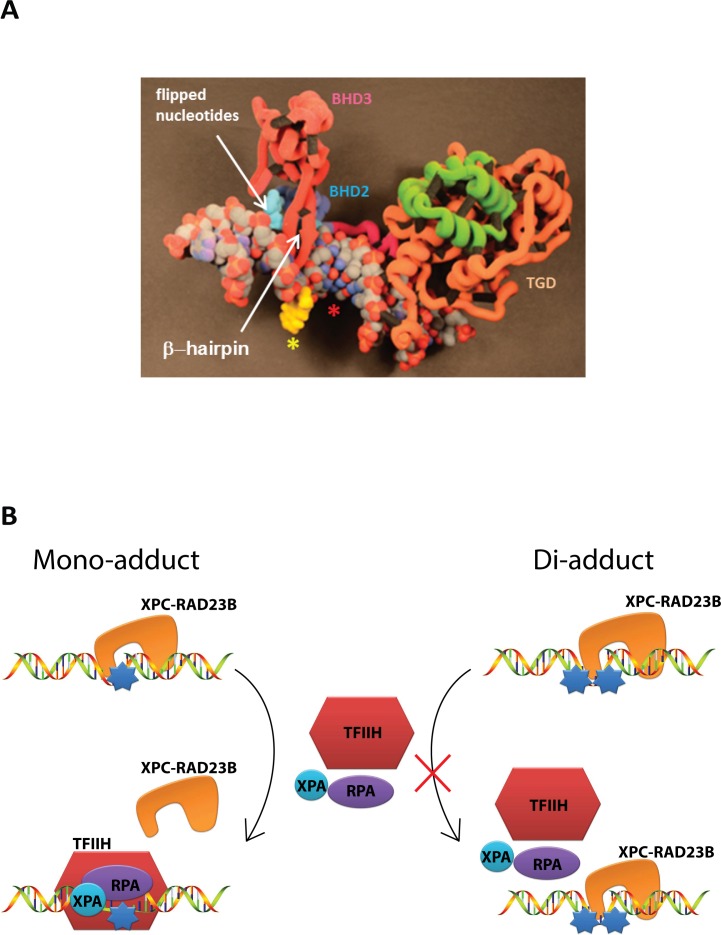
Fig 6. Proposed model for XPC interaction with DNA-adduct site.
(A) 3D-printed model (not simulated) to illustrate the potential binding of the yeast XPC-RAD23B ortholog, Rad4/Rad23, to the mono-G1-FAAF duplex based on PDB ID 2QSG. The β-hairpin domains (BHD2 and BHD3) and the transglutaminase-homology domain (TGD), which are involved in protein-DNA interaction, are indicated. The domains were adapted from previous crystal structure analysis by Min et al. [24]. The duplex sequence used in this model is identical with that of Min’s crystal work except that the CPD lesion was replaced by FAAF-G1 (yellow *, as shown). The site of additional FAAF in the di-G2G3-adduct is designated in red asterisk. The insertion of the BHD3 β-hairpin was accompanied with flipping of the mismatched bases (cyan) on the complimentary sequence. (B) A schematic illustrating the proposed mechanism of action where XPC is loosely bound to mono-FAAF adducted DNA (left) or tightly bound to di-FAAF adducted DNA (right). Following dissociation of XPC from the damage site subsequent NER factors are recruited to complete the excision of the damaged base; however, in the di-adduct situation XPC is retained on the damaged DNA, delaying successful NER completion.