Figure 1. Local nucleosome disassembly and reassembly around DSBs in human cells.
(A) Schematic demonstrating primer pairs spanning the I-PpoI site that were used to measure cutting and repair by quantitative PCR, while ChIP analysis was performed on the chromatin to the 5’ and 3’ of the I-PpoI site. (B) The kinetics of generation and repair of the DSB induced in the SLCO5A1 gene by the I-PpoI endonuclease. Shown is the average +/- SEM from three independent experiments. (C) Southern analysis of the kinetics of generation and repair of the DSB induced in the SLCO5A1 gene by the I-PpoI endonuclease from an independent time course from that shown in (B). (D) Quantitation of proportion of input DNA and H3 occupancy from ChIP analysis at 200, 500, 750 and 1000 bps (from left to right panels) away from the I-PpoI site within the SLCO5A1 gene. Shown is the average +/- SEM from three independent experiments. (E) (from top to bottom) DNA cutting analysis at the I-PpoI site within the RYR2 gene, input and H3 occupancy from H3 ChIP analysis 200bp from the I-PpoI site within the RYR2 gene. Shown is the average +/- SEM from three independent experiments.

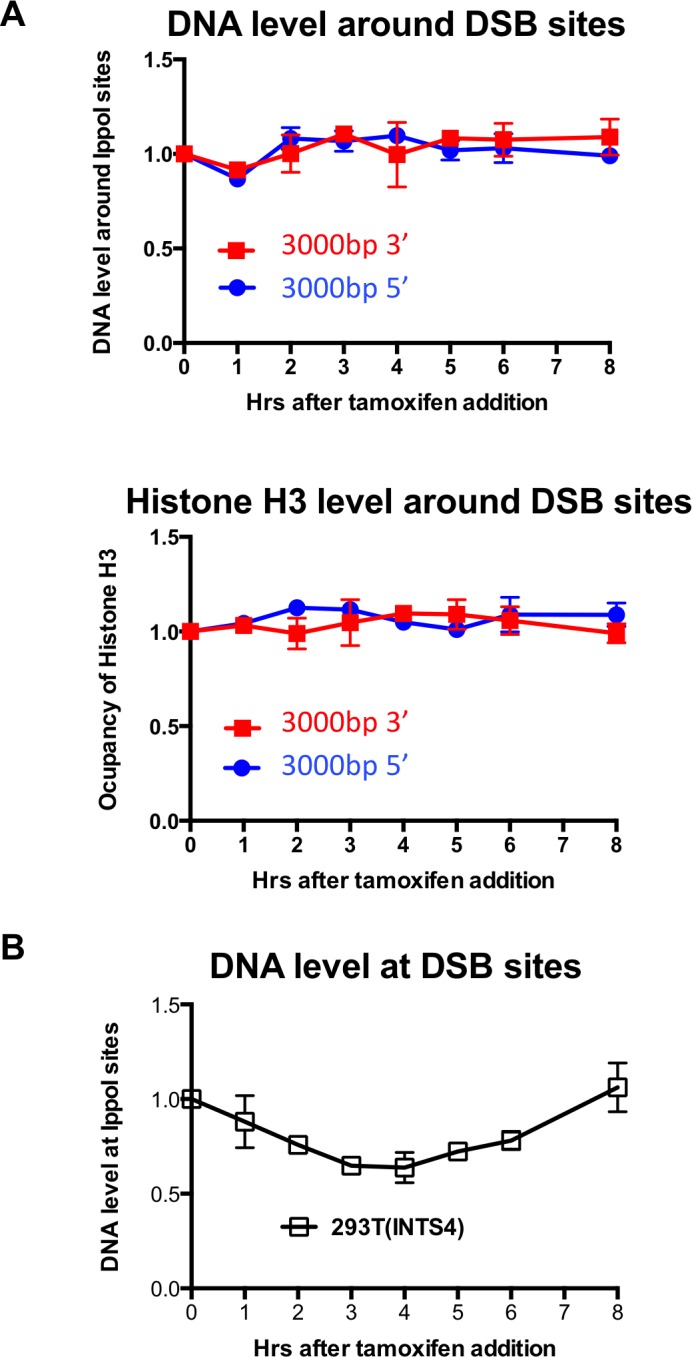
Figure 1—figure supplement 1. Limited H3 disassembly at a distance and I-PpoI cleavage at INTS4.