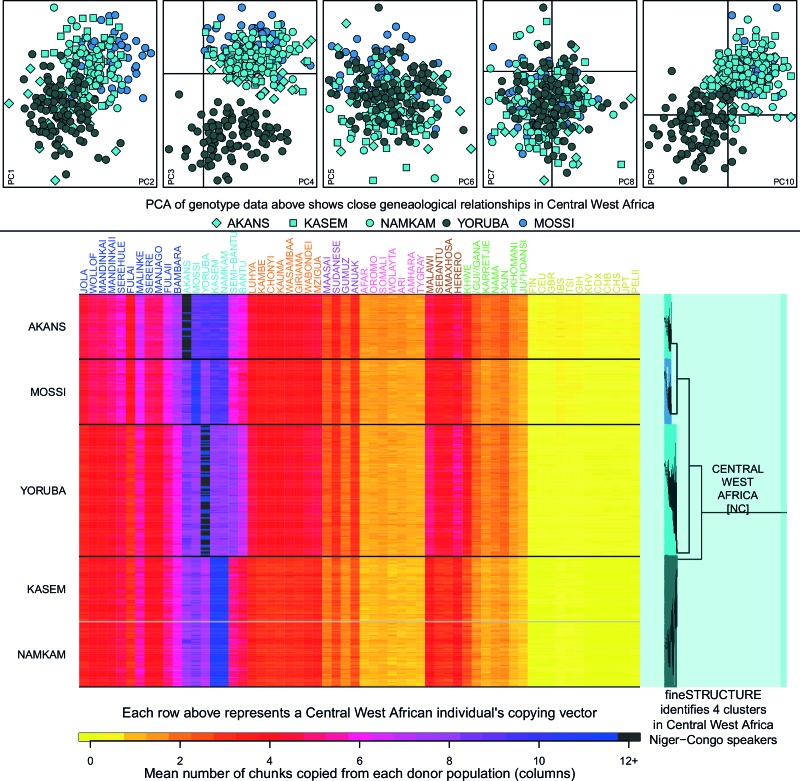
Here we show a comparison of principal components analysis (PCA), which uses genotype data, with fineSTRUCTURE, which uses haplotypic information in the form of painted chromosomes. The five plots in the top panel show the results of the main PCA based on genotype data. Symbols represent individuals and are detailed in the legend. PC3 differentiates the Yoruba from other groups in the region, but individuals from Central West Africa overlap at the remaining PCs, suggesting close genealogical relationships between individuals. The lower panel shows the results of the chromosome painting analysis, which we used with fineSTRUCTURE, where all individuals were allowed to copy from all other individuals. The rows of the heatmap represent un-normalised, individual copying vectors, with the mean number of chunks copied from each donor region as columns. Subtle differences in the copying of individuals from each of the five Central West African groups can be seen, which fineSTRUCTURE uses to cluster individuals into four clusters. We show a close-up of the fineSTRUCTURE tree from
Figure 1 on the right of the bottom panel. Each group separates into its own cluster, with the exception of two of the groups from Ghana, the Kasem and Namkam, which are put in the same fineSTRUCTURE cluster.