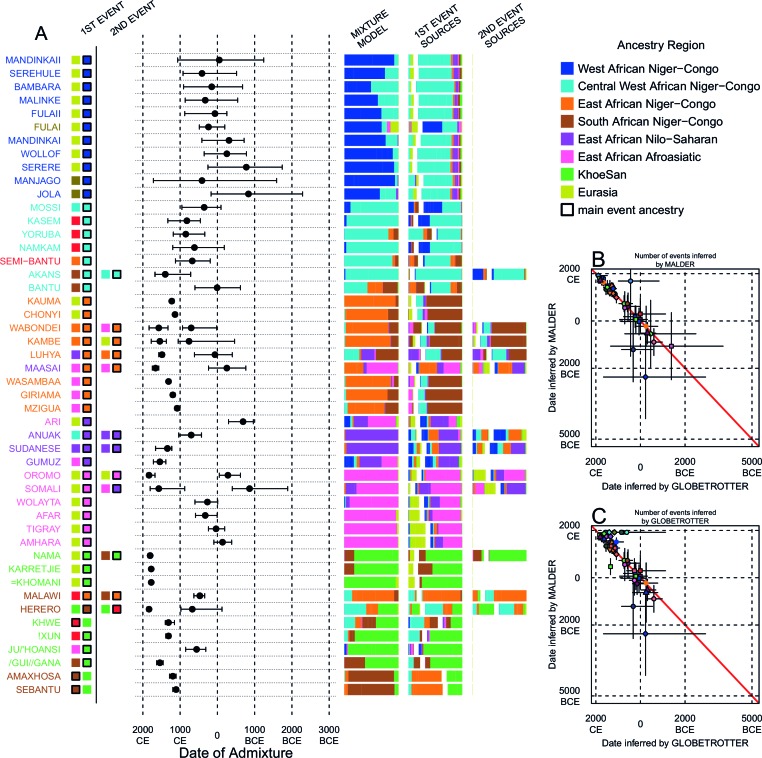
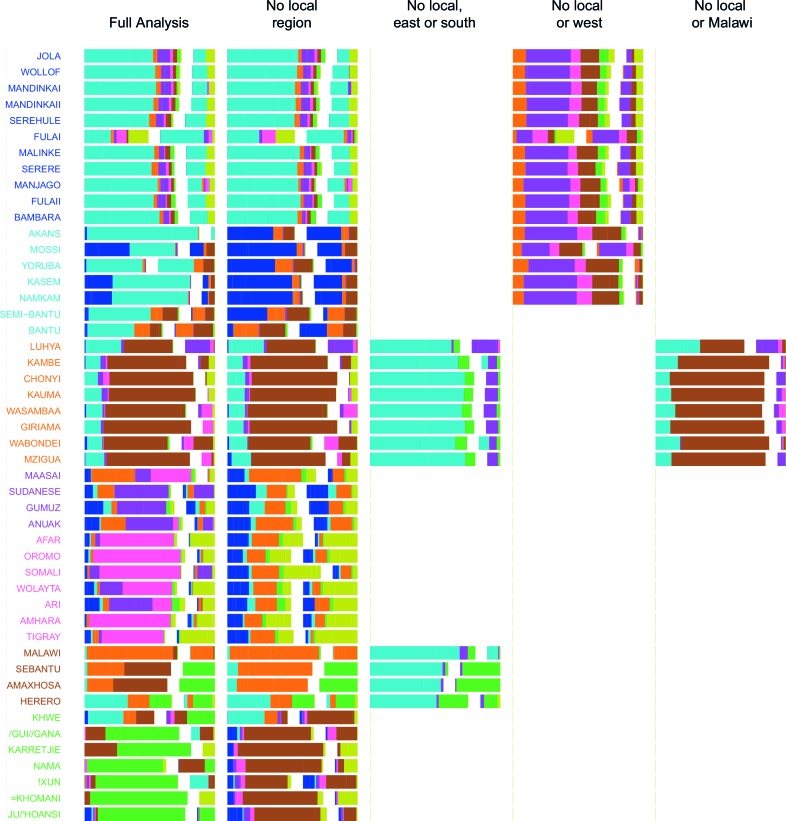
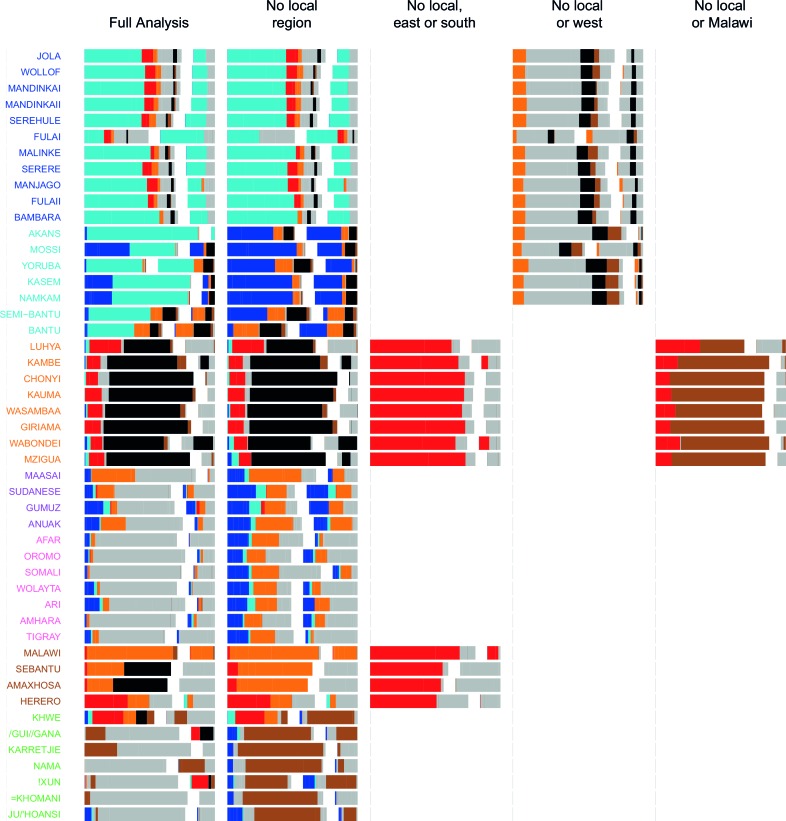
Figure 4. Inference of admixture in sub-Saharan African using GLOBETROTTER.
(A) For each group we show the ancestry region identity of the best matching source for the first and, if applicable, second events. Events involving sources that most closely match FULAI and SEMI-BANTU are highlighted by golden and red colours, respectively. Second events can be either multiway, in which case there is a single date estimate, or two-date in which case 2ND EVENT refers to the earlier event. The point estimate of the admixture date is shown as a black point, with 95% CI shown with lines. MIXTURE MODEL: We infer the ancestry composition of each African group by fitting its copying vector as a mixture of all other population copying vectors. The coefficients of this regression sum to 1 and are coloured by ancestry region. 1ST EVENT SOURCES and 2ND EVENT SOURCES show the ancestry breakdown of the admixture sources inferred by GLOBETROTTER, coloured by ancestry region as in the key top right. (B) and (C) Comparisons of dates inferred by MALDER and GLOBETROTTER. Because the two methods sometimes inferred different numbers of events, in (B) we show the comparison based on the inferred number of events in the MALDER analysis, and in (C) for the number of events inferred by GLOBETROTTER. Point symbols refer to populations and are as in Figure 1 and source data can be found in Figure 4—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.15266.026