Abstract
Transcription, the first step of gene expression, is exquisitely regulated in higher eukaryotes to ensure correct development and homeostasis. Traditional biochemical, genetic, and genomic approaches have proved successful at identifying factors, regulatory sequences, and potential pathways that modulate transcription. However, they typically only provide snapshots or population averages of the highly dynamic, stochastic biochemical processes involved in transcriptional regulation. Single molecule live-cell imaging has, therefore, emerged as a complementary approach capable of circumventing these limitations. By observing sequences of molecular events in real time as they occur in their native context, imaging has the power to derive cause-and-effect relationships and quantitative kinetics to build predictive models of transcription. Ongoing progress in fluorescence imaging technology has brought new microscopes and labeling technologies that now make it possible to visualize and quantify the transcription process with single-molecule resolution in living cells and animals. Here we provide an overview of the evolution and current state of transcription imaging technologies. We discuss some of the important concepts they uncovered and present possible future developments that might solve long-standing questions in transcriptional regulation.
Transcriptional regulation can be measured, described, and understood at several levels. Because much of the transcription reaction can be reconstituted in vitro from purified components, some aspects of its regulation can be addressed at the molecular level (Lemon et al. 2001; Fong et al. 2011, 2014). The structures of RNA polymerase II (Cramer et al. 2000; Gnatt et al. 2001) and many components of the preinitiation complex have been resolved at atomic resolution. Mechanistically how the basic reaction driving transcription works is therefore relatively well understood. Transcription has also been extensively studied at the cellular level using sophisticated genomic approaches such as ChIP-seq/exo and RNAseq (Ozsolak and Milos 2011; Shapiro et al. 2013; van Dijk et al. 2014) that can monitor the occupancy of polymerases and transcription factors (TFs) as well as RNA output on a genome-wide scale. These high-throughput studies provide one measure of gene expression output but often fail to reveal the underlying molecular mechanisms governing the exquisite regulation of transcription due to population averaging. Although in vitro studies have produced much of our knowledge regarding gene regulatory mechanisms, it is clear that genes are not regulated independently of each other. Likewise, genomic approaches are hampered by the inherent complexity of gene regulatory networks and the challenges posed by different stochastic processes involved in transcriptional control (Eldar and Elowitz 2010; Singer et al. 2014; Lin et al. 2015; Semrau and van Oudenaarden 2015).
Another complication is that at the single-cell nucleus level, DNA and chromatin are not randomly distributed in the nucleoplasm (Misteli 2007; Cremer and Cremer 2010; Dixon et al. 2012; Eagen et al. 2015). Instead, there is a spatially ordered hierarchy of nuclear structures, as well as highly dynamic transactions occurring between different genes, enhancers, and the protein TFs regulating them. The fundamental principles governing the interplay between nuclear organization and transcriptional regulation in vivo remain poorly understood.
The tracking of fluorescently tagged components of the transcription machinery in living or fixed single cells has begun to provide new insights into how the biochemistry of gene regulation operates within cells for a number of key regulatory molecules (Kusumi et al. 2014). Here we will review how different imaging techniques measuring TF dynamics along with nascent RNA production helped inform and change our understanding of gene regulation.
IMAGING APPROACHES TO MEASURE TF DYNAMICS AND GENOME ORGANIZATION
Observation of TF dynamics inside living cells is fundamental to a quantitative understanding of how precise spatiotemporal gene regulation is generated during animal development. Imaging modalities such as FRAP (fluorescence recovery after photobleaching), FCS (fluorescence correlation spectroscopy), SIM (structured illumination microscopy), and single-particle tracking (SPT) each provide unique advantages for measuring TF dynamics (Liu et al. 2015). For example, FRAP is an optical technique capable of quantifying the molecular diffusion and binding residence times in single cells (for review, see McNally 2008; Mueller et al. 2013). In combination with multiphoton microscopy of Drosophila polytene chromosomes, FRAP was used to study how the dynamics of TFs change at a discrete target gene during the heat shock response (Yao et al. 2006). FRAP can probe residence times of TFs on scales ranging from seconds (transcriptional activators and chromatin remodelers [McNally et al. 2000; Becker et al. 2002; Johnson et al. 2008]) to minutes (H1 [Misteli et al. 2000; Stasevich et al. 2010]) to hours (histones; for review, see Kimura 2005). At short timescales (milliseconds to seconds), FRAP measurement interpretations are model-dependent and could be biased by limited imaging acquisition speed, subdiffusion, and population averaging effect. However, FRAP has advantages over single-molecule imaging in detecting stable binding events that last for minutes and hours. This is because, for single-molecule imaging, fluorescently labeled TF molecules have to be excited by relatively high laser powers to generate enough photons for accurate localization, leading to the rapid expenditure of photon budget that in turn limits the temporal length scale of residence time measurements. Populationbased FRAP measurements require far less illumination powers and thus can be extended into much longer timescales.
FCS is a method capable of measuring absolute molecular concentrations, diffusion rates, and molecular interaction dynamics in live cells (Magde et al. 1972). The measurement is based on observation of a single molecule or several molecules within a diffraction-limited spot in solution or in a living cell. Although FCS measurements suffer from population averaging effects, FCS has advantages that complement SPT measurements. First, the observed molecules are continuously replenished by diffusion into the spatially restricted excitation volume. Thus, FCS allows observation for longer durations and does not require selection of specific molecules for observation. Second, because FCS measurements rely on fluorescence fluctuations rather than localization, fast detection paradigms can be used that make the technique sensitive enough to measure diffusion events on a much faster scale than is possible by SPT (Mazza et al. 2012; Chen et al. 2014b). By fitting FCS data to models of diffusion, one can assess the partition of nuclear factors between chromatin-bound and -free states. This technique is therefore very powerful at testing biophysical models of nuclear exploration—for example, addressing how TFs find their genomic targets and the importance of weak interactions in transcription regulation (Mueller et al. 2013). One important question is whether weak, nonproductive interactions between TFs and chromatin play a role in regulating TF availability and activity.
Decoding the complex behavior of single molecules enables measuring TF diffusion and TF:chromatin binding kinetics at this fundamental level that is often veiled in ensemble experiments. With recent advances in molecular imaging and chemical dyes (for review, see Liu et al. 2015), it has become possible to perform SPT of individual protein molecules in single live cells (Fig. 1A – C; Elf et al. 2007; Hager et al. 2009; Mazza et al. 2012; Abrahamsson et al. 2013; Gebhardt et al. 2013; Mueller et al. 2013; Chen et al. 2014b; Izeddin et al. 2014; Normanno et al. 2015). These rapidly emerging fast superresolution imaging platforms provide a means for visualizing and measuring the in vivo behavior of dynamically regulated TF-binding events at cis-regulatory DNA targets such as enhancers and core promoters. Once a DNA-binding protein reaches its target site, it becomes possible to estimate how long the protein stays bound to execute the desired outcomes such as transcriptional activation, chromatin remodeling, and even genome editing (Voss et al. 2011; Chen et al. 2014b; Knight et al. 2015; Normanno et al. 2015). These experiments have showed that eukaryotic TFs spend a substantial portion of their time freely diffusing (80%) (Chen et al. 2014b; Normanno et al. 2015). This behavior contrasts with prokaryotic TFs that typically spend most of their time associated with DNA (90%) (Elf et al. 2007), pointing to the important role that chromatin and DNA accessibility play in generating complexity in eukaryotic gene expression. Interestingly, distinct TFs have evolved different mobility properties: Although some factors explore the nucleus globally, others become confined to small subregions (Izeddin et al. 2014; for review, see Woringer et al. 2014). Finally, growing evidence indicates that spatial clustering is an important mode of regulation, either stably (minutes) in the case of TFs (Liu et al. 2014) or more dynamically (seconds) for the polymerase machinery (Cisse et al. 2013). Together, these findings show the importance of resolving the trajectories of biological factors in the complex space of the nucleus to address how robust regulation patterns can emerge from the stochastic behaviors of individual molecules.
Figure 1.
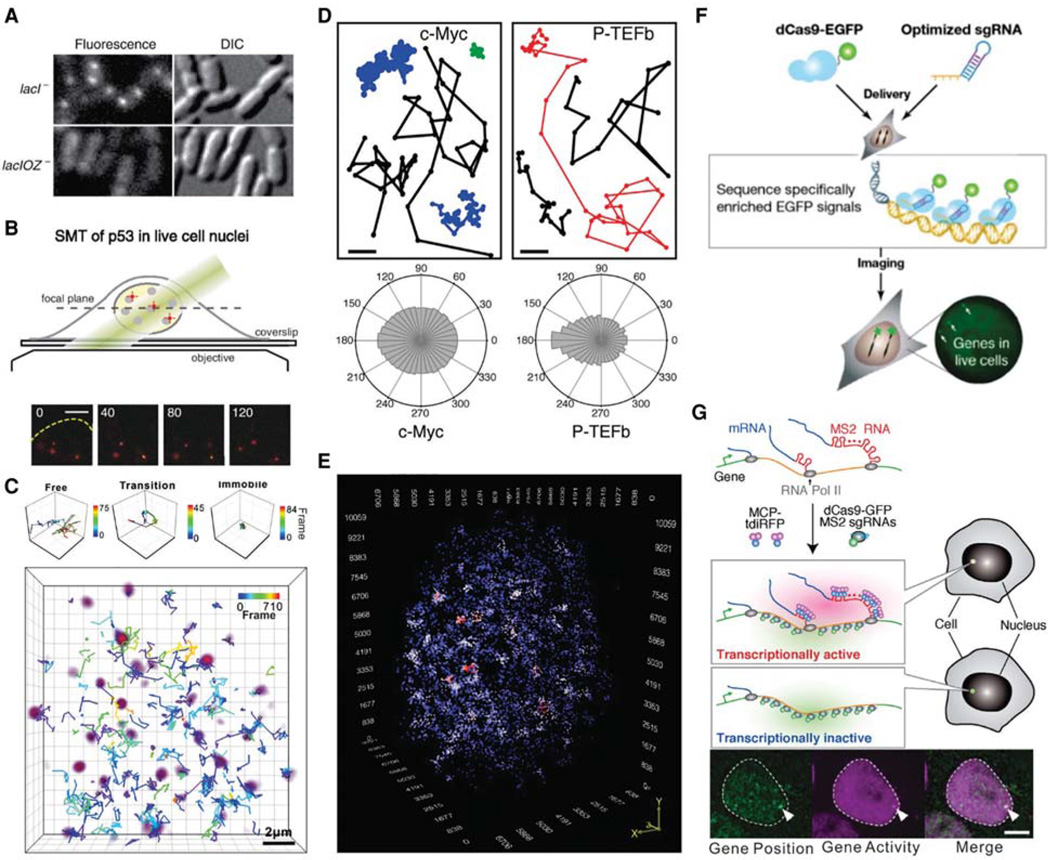
Live-cell imaging of TF dynamics and genome organization. (A) Detection of binding of the LacI factor to its cognate DNA sequence (LacO) in bacteria at the single-molecule level. Fluorescence (left) and DIC (right) images (1 sec exposure) of LacI-Venus expressed from plasmid in the lacI2 and lacIOZ2 strains, respectively. Fluorescent spots represent individual molecules of LacI. (B) 2D imaging of p53 dynamics in cancer cells. (Top) Optical setup. (Bottom) Individual HaloTag-p53 molecules (red spots) labeled with tetramethylrhodamine (TMR) were imaged at different time points (in microseconds; yellow dashed line indicates the nuclear boundary). (C) Tracking of individual TF in 3D. A HaloTag-Sox2 construct was expressed in a single embryonic stem cell, labeled with TMR and imaged using multifocus microscopy (MFM). Volume rendering of a Sox2 single-molecule image (purple) superimposed with single-molecule trajectories. Three molecules with distinct behaviors were selectively displayed on the top (from left to right, freely diffusing particle, particle undergoing a free/bound transition, and immobile molecule). Color bar shows the corresponding frame number. Scale bar, 2 mm. (D) Distinct TFs explore the nucleus differently. Examples of single-molecule traces for c-Myc and P-TEFb (top). Distribution histograms, in polar coordinates, of the angle u formed between the vectors of two consecutive translocation steps for c-Myc and P-TEFb. c-Myc angle distribution is isotropic, characteristic of a global explorer, whereas P-TEFb is a local explorer that reaches a genomic target in a position-dependent manner. (E) 3D organization of Sox2-binding sites in live mES cells. mES cells expressing HaloTag-Sox2 were labeled by the membrane-permeable JF549 dye and imaged on a lattice light-sheet microscope (300-nm z-steps; 50 msec per frame). Single molecules were localized and tracked in 3D. Sox2 molecules dwelling at the same position for .3 sec were counted as stably bound. Spots represent stably bound events; color codes the local density of stable localizations (blue, low density; red, high density). (F) Cas9-based live-cell gene locus labeling. Naturally repetitive and unique endogenous gene loci can be labeled based on sequence complementarity using fluorescent, nuclease-deficient Cas9 complexes (dCas-EGFP). (G) Simultaneous imaging of gene position and activity (ROLEX) in mES cells. A cassette of 24 tandem MS2 sequence was knocked into the Nanog gene and used as a target for both Cas9-based labeling of the DNA locus position (with dCas-GFP, green) and MS2 labeling to report on transcription activity (see text; MCP-tdiRFP, purple). (A, Reprinted from Elf et al. 2007, with permission from AAAS; B, reprinted from Mazza et al. 2012; C, adapted from Chen et al. 2014b; D, adapted from Izeddin et al. 2014; E, reprinted from Liu et al. 2014; F, reprinted from Chen et al. 2013, with permission from Elsevier; G, adapted from Ochiai et al. 2015.)
Combinatorial cis-regulatory networks encoded in animal genomes represent the foundational gene expression mechanism for directing cell-fate commitment and maintenance of cell identity by TFs. Currently, we have gained significant insights into static “snapshots” of genome organization by using two orthogonal fixed-cell-based approaches—chromatin conformation capture (3C) and microscopy techniques such as fluorescence in situ hybridization (FISH) (Misteli 2007; Bickmore 2013; Dekker et al. 2013; Liu 2015; Shachar et al. 2015). However, these techniques will not be able to provide detailed information about the kinetics and dynamics of the 3D genome as it operates in living cells. Recent advances in fluorescence microscopy and protein-labeling chemistry have advanced the possibility of mapping 3D TF-binding site organization in the nucleus of living mammalian cells and elucidating the local target search pattern and efficiency of TFs in finding and binding specific target sites in the highly heterogeneous subnuclear environment (Fig. 1D,E; Izeddin et al. 2014; Liu et al. 2014; Knight et al. 2015) as well as chromatin organization (Recamier et al. 2014). Experiments in bacteria have shown that live-cell imaging is able to monitor the looping of a DNA locus in real time (Hensel et al. 2013). Hopefully, with the further development of genome editing, live-cell mRNA/ locus labeling, and superresolution fluorescence microscopy techniques (Fig. 1F,G; Chen et al. 2013; Liu et al. 2015; Ochiai et al. 2015), it will soon become possible to use multicolor imaging experiments to report on the gene position, gene expression kinetics, and genome organization at the same time in single live eukaryotic cells. It is likely that both liveand fixed-cell techniques may need to be combined to decipher how the genome is structurally organized and dynamically functions in the nucleus (4D nucleome).
IMAGING SITES OF NASCENT TRANSCRIPTION
An important advantage of imaging is that although it is now routine to image and count absolute numbers of mRNA molecules in single cells using single-molecule mRNA FISH (Femino et al. 1998; Raj et al. 2008), reporters such as the MS2 and PP7 systems now make it possible to follow transcription in the native context of the living cells. These imaging modalities also provide an important qualitative difference from sequencing-based techniques, which require extracting DNA and RNA postmortem. These imaging reporters have made it possible to track transcription in various systems. Although transcription imaging was initially restricted to large, artificial gene arrays that generate bright signals (for review, see Darzacq et al. 2009), recent progress in microscopy has made it possible to follow in real time the expression of single genes in various systems, including endogenous loci in yeast (Larson et al. 2011), mammalian cells and live tissue (Lionnet et al. 2011a; Park et al. 2014), zebrafish (Campbell et al. 2015), or developing fly embryos (Garcia et al. 2013; Lucas et al. 2013; Bothma et al. 2014; Junker and van Oudenaarden 2014).
Imaging transcription in live cells or animals (e.g., using the MS2 system) can be done with frame rates of seconds or less. Such nascent mRNA imaging experiments have provided the direct demonstration of the biological process called transcriptional bursting: Many genes tend to be transcribed intermittently, producing a large number of transcripts in short pulses interspersed by long, silent intervals (Chubb et al. 2006; Lionnet and Singer 2012). This finding has fundamental consequences for gene expression, as this type of expression pattern typically yields high cell-to-cell variation, which cell populations and tissues then need to either limit or use to their benefit (Balazsi et al. 2011; Little et al. 2013). Although topological constraints on the genomic DNA were recently shown to explain bursting in a bacterial gene model (Chong et al. 2014), the generality of this mechanism is unclear and nascent transcription imaging techniques will likely play an important role in providing a molecular basis for this phenomenon. Importantly, imaging techniques are crucial to address how gene networks evolved harnessing these dynamics to produce highly regulated biological processes such as development (Bothma et al. 2014) or stress response, where the relative timing of transcription pulses proves crucial for adequate expression of target genes (Lin et al. 2015).
The high temporal resolutions accessible through imaging also make it possible to quantitatively measure in live cells not only transcriptional outputs but also kinetic rates of substeps within the transcription process itself, a prerequisite to determine the regulatory checkpoints during transcription. Such experiments were initially performed using FRAP, in which the kinetics of fluorescence recovery after photobleaching the nascent mRNA signal enabled extracting transcription rates (Darzacq et al. 2007). FRAP approaches are best performed on artificial cell lines harboring large arrays of repeats of a reporter gene. More recently, high-resolution imaging coupled to sophisticated correlation techniques have allowed quantifying initiation, elongation, and splicing rates from nascent mRNA imaging experiments (Larson et al. 2011; Lionnet et al. 2011b; Martin et al. 2013; Coulon et al. 2014).
IMAGING VERSUS GENOMIC APPROACHES
Genomics techniques are moving at a fast pace (Junker and van Oudenaarden 2014), and it is now possible in one experiment to obtain the genome-wide expression profiles of thousands of individual cells. However, imaging techniques still hold a large number of advantages. Light microscopy has exquisite resolution, in both space and time. Transcription imaging is by nature a single-cell technique, and it allows identifying rare relevant cell types in complex mixed cell populations or tissues and measuring their expression quantitatively (Cochella and Hobert 2012). In contrast with genomics-based single-cell techniques, transcription imaging can easily be combined with other observables. This allows correlating transcription levels to important variables such as upstream TF availability or other factors (e.g., cell size or cell-cycle stage) that are usually hard to access in genomics experiments. Applying techniques used in superresolution, it is currently possible to image individual molecules with a localization precision of 40 nm in x,y (a cube of 40-nm size encompasses on average a few kilobase pairs of genomic DNA in a typical mammalian nucleus). This spatial resolving power allows comparing the expression of individual alleles within the same nucleus to evaluate their correlation, an important tool to study transcription bursting and expression variability (Gandhi et al. 2011; Kalo et al. 2015). Although constitutively expressed genes are transcribed stochastically in an uncorrelated fashion, activation of a signaling pathway can trigger a correlated behavior (Kalo et al. 2015). The current spatial resolution is on par with the length scales relevant to address one of the long-standing questions in the transcription field: What is the molecular mechanism of enhancer – promoter communication? So far this question has been tackled only by ensemble techniques (3C and variants) or fixed-cell-based imaging (DNA FISH) where the preservation of fine chromatin structure after fixation, DNA denaturation, and permeabilization steps may be suspect. Live-cell imaging is currently the only technique that has the potential to measure both the transcription levels of a gene and its native chromatin architecture, at the single-locus level. By fluorescently labeling loci in cells with various tagging techniques (Chen et al. 2013; Hensel et al. 2013), one could imagine correlating, in real time, the looping of an enhancer with its cognate promoter and correlate it with the transcriptional activity of the regulated gene. Such experiments will be crucial to help understand how chromatin architecture and TF binding regulate the expression of developmental genes.
Finally, throughput has long been a limitation of imaging-based techniques. However, recent progress in barcoding in situ techniques and in situ sequencing (Levsky et al. 2002; Lubeck and Cai 2012; Levesque and Raj 2013; Lee et al. 2014; Lubeck et al. 2014; Chen et al. 2015) are now making it possible to image tens to thousands of genes in the same cell at the same time, opening the door to studying network-level regulation in single cells.
THE FUTURE OF IMAGING TRANSCRIPTION
We are on the verge of some profound insights into how transcription occurs. We now have tools to investigate this and some yet to be perfected. The ability to create labeled genes and TFs provides an entrée into the earliest events preceding transcription. New microscope developments can image these events at the millisecond timescale and, ultimately, in the unperturbed cells of the body.
What a cell is, and will become, is the sum of thousands of probabilistic events that, integrated over time, form a more deterministic transcriptome and ultimately the proteome of a cell. The TFs that drive these genes are themselves products of processes that were initiated by master regulators that originated in the early ancestors of the cell, the fertilized oocyte. A cascade begins and wave after wave of factors flood through the nucleus, opening up new areas of chromatin for expression. Imaging has an important role to play in addressing the long-standing question of the mechanisms regulating the interplay between the spatial organization of the nucleus and transcription activity during development. Currently, in fixed cells, it is possible to image the activity of many genes at a moment in time by hybridizing fluorescent probes to their nascent chains (Femino et al. 1998, 2003; Levsky et al. 2002, 2007). An active gene generates many nascent chains by virtue of the many elongating polymerases. By using repetitive hybridizations and a spectral barcode, it is theoretically possible to interrogate every gene in the human genome at a particular time point (Levsky et al. 2002; Lubeck and Cai 2012; Lubeck et al. 2014). Importantly this can be done while preserving the spatial information in the nucleus, thereby creating a “star map” or constellation of genes with their sequence identities. Theoretically, the entire human transcriptome could be imaged within one nucleus, although only a small subset of genes would be expected to be active at any one moment. The resolution of the microscope, even in the z-axis, is sufficient to detect around 25,000 diffraction-limited spots in the average mammalian nucleus, even without superresolution (based on a radius of 5 mm and a resolution of 200 nm in x,y and 500 nm in z). Such techniques will be able to connect observations of clustering in live cells (Cisse et al. 2013; Liu et al. 2014) to underlying organizational principles and to assess their functional relevance. Specifically, it will be important to assess the role of enhancers as tethered transcriptional activators working from hundreds of kilobase pairs away (Deng et al. 2012, 2014; Liu et al. 2014).
Although the snapshot in time provides valuable information about spatial relationships, the dynamics are missing and hence so is any understanding about mechanistic events leading up to gene activation. It is not yet possible to uniquely tag a large number of genes for in vivo imaging, but even several well-chosen ones will be informative (Larson et al. 2011; Lionnet et al. 2011a; Park et al. 2014; Ma et al. 2015). Although labeling genes and factors at endogenous loci in higher eukaryotes has long been a challenging proposal, the advent of CRISPR/ Cas9 technologies is enabling almost unrestricted access to reporter lines and animals. The construction of microscopes that can simultaneously excite many fluorophores and image in each color simultaneously on highly registered cameras will allow the correlation of many desired factors: genes by labeling using MBS or PBS stem loops, TFs, or histone modifiers (Grunwald and Singer 2010; Stasevich et al. 2014; Monnier et al. 2015). This would allow the ordering of events (histone modification, TF binding, complex formation, polymerase initiation, nascent chain formation, splicing, and termination), and all could conceivably be done on the same cell with spectral identification using multiple fluors coupled with variants of halo tags or Cas9 or proteins yet to be discovered that identify specific RNA or DNA sequences (Zhang et al. 2014; Grimm et al. 2015; Liu et al. 2015). For instance, the current methods for imaging genes and RNA are rather cumbersome, because of the use of repetitive sequences for increasing the signal to noise. Specialized complexes such as Cas9 coupled to a guide RNA can recognize specific DNA or RNA sequences (Chen et al. 2013; O’Connell et al. 2014; Ma et al. 2015). However, they do not currently generate enough signal at their target to be detected above background. Spatial frequency filtering methods, however, may allow the detection of single proteins that dock at their target site and hence “pull-out” the signal from the diffusing background (Cisse et al. 2013; Normanno et al. 2015). The fast pace of microscopy technology development (for review, see Liu et al. 2015) is likely to bring new imaging modalities to explore these questions with higher signal-to-noise ratios. For instance, SIM increases the spatial resolution twofold compared with wide-field imaging, and requires moderate light intensities. The combination of live-cell SIM with single-molecule imaging modalities could be a powerful tool to study TF diffusion and chromatin-binding dynamics in the context of subcellular structures that regulate these events. Other imaging modalities, such as light sheet microscopy (Chen et al. 2014a), FRET (Forster resonance energy transfer), FLIM (fluorescence lifetime imaging microscopy), and polarization optics, are also poised to make greater contributions to probing transcription in live cells by multiplexing with rapidly advancing imaging and labeling technologies.
The ultimate goal in studying transcription is to see it in action in cells within the living and developing animal. The advances made on single cells in culture, under optimal optical conditions, will need to be transferred to the living animal where imaging is much more difficult. Only then will we understand the mechanisms controlling development and homeostasis. The suboptimal conditions of imaging in animals involve the light scattering and penetration problems inherent in tissue. What is required is the ability to circumvent these problems. There are four main approaches for this: the sample chosen, the imaging modality, the detectable reagents, and the processing of the images. Currently, the most effective means of penetrating using photons is the two-photon microscope, where a pulsed laser delivers lower-energy photons to a focused spot to excite a fluorochrome when they arrive nearly simultaneously and sum (Denk et al. 1990). Therefore, two photons at 960-nm wavelength will excite a fluorophore with an excitation peak at 480 nm. Because tissue is more transparent at 960 nm, and because the photons do not excite any fluorophores except at the point of focus where the flux is highest, photodamage, bleaching, and the activation of out-of-focus fluors are obviated. This makes the signal more visible relative to the background. However, at 480 nm, the emitted light causes autofluorescence and lower penetration, so newer red fluorophores with a two-photon cross section can emit at longer wavelengths, and hence their signal is less susceptible to scattering. Another method of imaging animals is by use of a light sheet that illuminates a thin, planar section of tissue (Keller et al. 2008). The image is collected perpendicular to this, so only the illuminated section is visible. Alternatively, a very thin beam of light can be scanned across the sample, allowing even less total illumination during imaging, and capable of detecting single molecules (Planchon et al. 2011). In the latest advance, lattice light sheet microscopy, a lattice of light illuminates the sample to create sectioning down to few hundreds of nanometers (Chen et al. 2014a). For these experiments, an animal on the scale of the microscopic range is desirable, such as zebrafish, which is more optically transparent, or Drosophila early embryos (Lagha et al. 2012; Bothma et al. 2014). Finally, recent developments in aberration compensation such as adaptive optics can correct the distortions in images collected from deep inside tissues (Wang et al. 2015). This allows for greater depth penetration and higher resolution. Ultimately it may be possible to use other modalities than photonbased imaging. Reagents that report position information on the larger scale, such as MRI or PET, could provide gene expression information on proteins that can act as contrast reagents, such as those that sequester iron (Wu et al. 2014). Although the resolution is not subcellular, the overall position of the expression within the living organism may be an important means to understand developmental or disease processes.
Ultimately everything biological depends on gene expression and the more ways we can develop to see the details, the more we will understand about how cells respond to their native environment.
Acknowledgments
This work is supported by a grant from the 4D Nucleome, U01 EB021236 to R.A.C., X.D., R.H.S., and T.L.
REFERENCES
- Abrahamsson S, Chen J, Hajj B, Stallinga S, Katsov AY, Wisniewski J, Mizuguchi G, Soule P, Mueller F, Dugast Darzacq C, et al. Fast multicolor 3D imaging using aberrationcorrected multifocus microscopy. Nat Methods. 2013;10:60–63. doi: 10.1038/nmeth.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazsi G, van Oudenaarden A, Collins JJ. Cellular decision making and biological noise: From microbes to mammals. Cell. 2011;144:910–925. doi: 10.1016/j.cell.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Baumann C, John S, Walker DA, Vigneron M, McNally JG, Hager GL. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–1194. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA. The spatial organization of the human genome. Annu Rev Genomics Hum Genet. 2013;14:67–84. doi: 10.1146/annurev-genom-091212-153515. [DOI] [PubMed] [Google Scholar]
- Bothma JP, Garcia HG, Esposito E, Schlissel G, Gregor T, Levine M. Dynamic regulation of eve stripe 2 expression reveals transcriptional bursts in living Drosophila embryos. Proc Natl Acad Sci. 2014;111:10598–10603. doi: 10.1073/pnas.1410022111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PD, Chao JA, Singer RH, Marlow FL. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development. 2015;142:1368–1374. doi: 10.1242/dev.118968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA, III, Liu Z, et al. Lattice light-sheet microscopy: Imaging molecules to embryos at high spatiotemporal resolution. Science. 2014a;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, et al. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014b;156:1274–1285. doi: 10.1016/j.cell.2014.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S, Chen C, Ge H, Xie XS. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. Real-time dynamics of RNA polymerase II clustering in live human cells. Science. 2013;341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- Cochella L, Hobert O. Embryonic priming of a miRNA locus predetermines postmitotic neuronal left/right asymmetry in C. elegans. Cell. 2012;151:1229–1242. doi: 10.1016/j.cell.2012.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Ferguson ML, de Turris V, Palangat M, Chow CC, Larson DR. Kinetic competition during the transcription cycle results in stochastic RNA processing. ELife. 2014;3 doi: 10.7554/eLife.03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Fu J, Gnatt AL, Maier-Davis B, Thompson NE, Burgess RR, Edwards AM, David PR, Kornberg RD. Architecture of RNA polymerase II and implications for the transcription mechanism. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Shav-Tal Y, de Turris V, Brody Y, Shenoy SM, Phair RD, Singer RH. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, et al. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nat Rev Genet. 2013;14:390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell. 2012;149:1233–1244. doi: 10.1016/j.cell.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Rupon JW, Krivega I, Breda L, Motta I, Jahn KS, Reik A, Gregory PD, Rivella S, Dean A, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagen KP, Hartl TA, Kornberg RD. Stable chromosome condensation revealed by chromosome conformation capture. Cell. 2015;163:934–946. doi: 10.1016/j.cell.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elf J, Li GW, Xie XS. Probing transcription factor dynamics at the single-molecule level in a living cell. Science. 2007;316:1191–1194. doi: 10.1126/science.1141967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Femino AM, Fogarty K, Lifshitz LM, Carrington W, Singer RH. Visualization of single molecules of mRNA in situ. Methods Enzymol. 2003;361:245–304. doi: 10.1016/s0076-6879(03)61015-3. [DOI] [PubMed] [Google Scholar]
- Fong YW, Inouye C, Yamaguchi T, Cattoglio C, Grubisic I, Tjian R. A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell. 2011;147:120–131. doi: 10.1016/j.cell.2011.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Ho JJ, Inouye C, Tjian R. The dyskerin ribonucleoprotein complex as an OCT4/SOX2 coactivator in embryonic stem cells. Elife. 2014;3:e03573. doi: 10.7554/eLife.03573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi SJ, Zenklusen D, Lionnet T, Singer RH. Transcription of functionally related constitutive genes is not coordinated. Nat Struct Mol Biol. 2011;18:27–34. doi: 10.1038/nsmb.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia HG, Tikhonov M, Lin A, Gregor T. Quantitative imaging of transcription in living Drosophila embryos links polymerase activity to patterning. Curr Biol. 2013;23:2140–2145. doi: 10.1016/j.cub.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt JC, Suter DM, Roy R, Zhao ZW, Chapman AR, Basu S, Maniatis T, Xie XS. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- Grimm JB, English BP, Chen J, Slaughter JP, Zhang Z, Revyakin A, Patel R, Macklin JJ, Normanno D, Singer RH, et al. A general method to improve fluorophores for livecell and single-molecule microscopy. Nat Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald D, Singer RH. In vivo imaging of labelled endogenous b-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell. 2009;35:741–753. doi: 10.1016/j.molcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel Z, Weng X, Lagda AC, Xiao J. Transcription-factor-mediated DNA looping probed by high-resolution, singlemolecule imaging in live E. coli cells. PLoS Biol. 2013;11:e1001591. doi: 10.1371/journal.pbio.1001591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izeddin I, Recamier V, Bosanac L, Cisse II, Boudarene L, Dugast-Darzacq C, Proux F, Benichou O, Voituriez R, Bensaude O, et al. Single-molecule tracking in live cells reveals distinct target-search strategies of transcription factors in the nucleus. ELife. 2014;3 doi: 10.7554/eLife.02230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TA, Elbi C, Parekh BS, Hager GL, John S. Chromatin remodeling complexes interact dynamically with a glucocorticoid receptor-regulated promoter. Mol Biol Cell. 2008;19:3308–3322. doi: 10.1091/mbc.E08-02-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junker JP, van Oudenaarden A. Every cell is special: Genome-wide studies add a new dimension to single-cell biology. Cell. 2014;157:8–11. doi: 10.1016/j.cell.2014.02.010. [DOI] [PubMed] [Google Scholar]
- Kalo A, Kanter I, Shraga A, Sheinberger J, Tzemach H, Kinor N, Singer RH, Lionnet T, Shav-Tal Y. Cellular levels of signaling factors are sensed by b-actin alleles to modulate transcriptional pulse intensity. Cell Rep. 2015;11:419–432. doi: 10.1016/j.celrep.2015.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- Kimura H. Histone dynamics in living cells revealed by photobleaching. DNA repair. 2005;4:939–950. doi: 10.1016/j.dnarep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Knight SC, Xie L, Deng W, Guglielmi B, Witkowsky LB, Bosanac L, Zhang ET, El Beheiry M, Masson JB, Dahan M, et al. Dynamics of CRISPR-Cas9 genome interrogation in living cells. Science. 2015;350:823–826. doi: 10.1126/science.aac6572. [DOI] [PubMed] [Google Scholar]
- Kusumi A, Tsunoyama TA, Hirosawa KM, Kasai RS, Fujiwara TK. Tracking single molecules at work in living cells. Nat Chem Biol. 2014;10:524–532. doi: 10.1038/nchembio.1558. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends Genet. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Zenklusen D, Wu B, Chao JA, Singer RH. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science. 2011;332:475–478. doi: 10.1126/science.1202142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SS, Li C, Amamoto R, et al. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414:924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- Levesque MJ, Raj A. Single-chromosome transcriptional profiling reveals chromosomal gene expression regulation. Nat Methods. 2013;10:246–248. doi: 10.1038/nmeth.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- Levsky JM, Shenoy SM, Chubb JR, Hall CB, Capodieci P, Singer RH. The spatial order of transcription in mammalian cells. J Cell Biochem. 2007;102:609–617. doi: 10.1002/jcb.21495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Sohn CH, Dalal CK, Cai L, Elowitz MB. Combinatorial gene regulation by modulation of relative pulse timing. Nature. 2015;527:54–58. doi: 10.1038/nature15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Singer RH. Transcription goes digital. EMBO Rep. 2012;13:313–321. doi: 10.1038/embor.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Methods. 2011a;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Wu B, Grunwald D, Singer RH, Larson DR. Nuclear physics: Quantitative single-cell approaches to nuclear organization and gene expression. Cold Spring Harb Symp Quant Biol. 2011b;75:113–126. doi: 10.1101/sqb.2010.75.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Tikhonov M, Gregor T. Precise developmental gene expression arises from globally stochastic transcriptional activity. Cell. 2013;154:789–800. doi: 10.1016/j.cell.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. Lighting up genes in single cells at scale. Cell. 2015;162:705–707. doi: 10.1016/j.cell.2015.07.052. [DOI] [PubMed] [Google Scholar]
- Liu Z, Legant WR, Chen BC, Li L, Grimm JB, Lavis LD, Betzig E, Tjian R. 3D imaging of Sox2 enhancer clusters in embryonic stem cells. ELife. 2014;3:e04236. doi: 10.7554/eLife.04236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Lavis LD, Betzig E. Imaging live-cell dynamics and structure at the single-molecule level. Mol Cell. 2015;58:644–659. doi: 10.1016/j.molcel.2015.02.033. [DOI] [PubMed] [Google Scholar]
- Lubeck E, Cai L. Single-cell systems biology by superresolution imaging and combinatorial labeling. Nat Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Single-cell in situ RNA profiling by sequential hybridization. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas T, Ferraro T, Roelens B, De Las Heras Chanes J, Walczak AM, Coppey M, Dostatni N. Live imaging of bicoiddependent transcription in Drosophila embryos. Curr Biol. 2013;23:2135–2139. doi: 10.1016/j.cub.2013.08.053. [DOI] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magde D, Webb WW, Elson E. Thermodynamic fluctuations in a reacting system—Measurement by fluorescence correlation spectroscopy. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- Martin RM, Rino J, Carvalho C, Kirchhausen T, Carmo-Fonseca M. Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. Cell Rep. 2013;4:1144–1155. doi: 10.1016/j.celrep.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazza D, Abernathy A, Golob N, Morisaki T, McNally JG. A benchmark for chromatin binding measurements in live cells. Nucleic Acids Res. 2012;40:e119. doi: 10.1093/nar/gks701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally JG. Quantitative FRAP in analysis of molecular binding dynamics in vivo. Methods Cell Biol. 2008;85:329–351. doi: 10.1016/S0091-679X(08)85014-5. [DOI] [PubMed] [Google Scholar]
- McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–881. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- Monnier N, Barry Z, Park HY, Su KC, Katz Z, English BP, Dey A, Pan K, Cheeseman IM, Singer RH, et al. Inferring transient particle transport dynamics in live cells. Nat Methods. 2015;12:838–840. doi: 10.1038/nmeth.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F, Stasevich TJ, Mazza D, McNally JG. Quantifying transcription factor kinetics: At work or at play? Crit Rev Biochem Mol Biol. 2013;48:492–514. doi: 10.3109/10409238.2013.833891. [DOI] [PubMed] [Google Scholar]
- Normanno D, Boudarene L, Dugast-Darzacq C, Chen J, Richter C, Proux F, Benichou O, Voituriez R, Darzacq X, Dahan M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat Commun. 2015;6:7357. doi: 10.1038/ncomms8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai H, Sugawara T, Yamamoto T. Simultaneous live imaging of the transcription and nuclear position of specific genes. Nucleic Acids Res. 2015;43:e127. doi: 10.1093/nar/gkv624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: Advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, LopezJones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science. 2014;343:422–424. doi: 10.1126/science.1239200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon TA, Gao L, Milkie DE, Davidson MW, Galbraith JA, Galbraith CG, Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recamier V, Izeddin I, Bosanac L, Dahan M, Proux F, Darzacq X. Single cell correlation fractal dimension of chromatin: A framework to interpret 3D single molecule super-resolution. Nucleus. 2014;5:75–84. doi: 10.4161/nucl.28227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau S, van Oudenaarden A. Studying lineage decision-making in vitro: Emerging concepts and novel tools. Annu Rev Cell Dev Biol. 2015;31:317–345. doi: 10.1146/annurev-cellbio-100814-125300. [DOI] [PubMed] [Google Scholar]
- Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. Identification of gene positioning factors using high-throughput imaging mapping. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- Singer ZS, Yong J, Tischler J, Hackett JA, Altinok A, Surani MA, Cai L, Elowitz MB. Dynamic heterogeneity and DNA methylation in embryonic stem cells. Mol Cell. 2014;55:319–331. doi: 10.1016/j.molcel.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, Mueller F, Brown DT, McNally JG. Dissecting the binding mechanism of the linker histone in live cells: An integrated FRAP analysis. EMBO J. 2010;29:1225–1234. doi: 10.1038/emboj.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, Tokunaga M, Nagase T, Nozaki N, McNally JG, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–275. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30:418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–554. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Sun W, Richie CT, Harvey BK, Betzig E, Ji N. Direct wavefront sensing for high-resolution in vivo imaging in scattering tissue. Nat Commun. 2015;6:7276. doi: 10.1038/ncomms8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woringer M, Darzacq X, Izeddin I. Geometry of the nucleus: A perspective on gene expression regulation. Curr Opin Chem Biology. 2014;20:112–119. doi: 10.1016/j.cbpa.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Wu B, Chen J, Singer RH. Background free imaging of single mRNAs in live cells using split fluorescent proteins. Sci Rep. 2014;4:3615. doi: 10.1038/srep03615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Munson KM, Webb WW, Lis JT. Dynamics of heat shock factor association with native gene loci in living cells. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Revyakin A, Grimm JB, Lavis LD, Tjian R. Single-molecule tracking of the transcription cycle by subsecond RNA detection. Elife. 2014;3:e01775. doi: 10.7554/eLife.01775. [DOI] [PMC free article] [PubMed] [Google Scholar]