Abstract
Pseudomonas aeruginosa and Staphylococcus aureus are versatile bacterial pathogens and common etiological agents in polymicrobial infections. Microbial communities containing both of these pathogens are shaped by interactions ranging from parasitic to mutualistic, with the net impact of these interactions in many cases resulting in enhanced virulence. Polymicrobial communities of these organisms are further defined by multiple aspects of the host environment, with important implications for disease progression and therapeutic outcomes. This mini-review highlights the impact of these interactions on the host and individual pathogens, the molecular mechanisms that underly these interactions, and host-specific factors that drive interactions between these two important pathogens.
Keywords: Pseudomonas aeruginosa, Staphylococcus aureus, cystic fibrosis, polymicrobial infections, alkylquinolones
INTRODUCTION
Polymicrobial communities are prevalent in many infectious disease states, including surgical and diabetic foot wounds, otitis media, oral infections, and cystic fibrosis (CF) lung disease (Peters et al. 2012). While the importance of these communities in infectious disease has long been understood, the advent of cutting-edge technologies is yielding new insights into the complexities and impacts of multi-species infections on human health (Marsland and Gollwitzer 2014; Surette 2013; Lynch and Bruce 2013; Price et al. 2013; Lipuma 2010; Rogers et al. 2010; Lyczak, Cannon, and Pier 2002; Filkins and O'Toole 2015). Despite these advances, the impact of polymicrobial interactions on infectious disease remains difficult to dissect in a laboratory environment. Contributing to the complexity of these studies is a dearth of appropriate models that accurately reflect the host environment. In particular, models for studying polymicrobial interactions must take into account the availability of required nutrients, impacts of host immunity factors, and changes in host physiology that occur as a result of infection and/or the underlying disease state.
Biological interactions in polymicrobial communities are most commonly defined based on the outcome of the interaction on each of the two participating species: parasitism refers to a relationship in which one organism benefits at the cost of the other, commensalism indicates that one organism benefits from the relationship with no effect on the other, and mutualism is defined as a relationship where both organisms benefit. Regardless of their outcome on the individual species, each of these interactions has the potential to accelerate disease progression when it occurs at the site of an infection. In these cases, the term “synergism” has been applied to denote cooperativity – i.e. enhanced virulence capacity during co-infection as compared to that of either microbe alone (Murray et al. 2014). In contrast, antagonism occurs when the presence of two or more species protects the host from disease that occurs when only one species is present. As will be discussed in this review, defining specific interactions as mutualistic or antagonistic can be subjective, as negative effects on growth rates of one species may promote survival of that species in specific host environments.
In this mini-review, we will provide an overview of interactions that have been described between Staphylococcus aureus and Pseudomonas aeruginosa, two versatile bacterial pathogens that commonly inhabit the CF lung and chronic wound infections (Stacy et al. 2016; Peters et al. 2012; Foundation 2014). Synergism between S. aureus and P. aeruginosa has been observed in multiple models of infection, including wounds and chronic lung infection (Sibley et al. 2008; Korgaonkar et al. 2013; Korgaonkar and Whiteley 2011; DeLeon et al. 2014; Dalton et al. 2011). Numerous studies have revealed both mutualistic and parasitic interactions that drive the synergistic impact of these two pathogens on infectious disease progression. We will specifically discuss the molecular mechanisms guiding these interactions, as well as their implications for the progression and outcome of polymicrobial infections.
COMMENSAL AND MUTUALISTIC INTERACTIONS
Both S. aureus and P. aeruginosa exhibit intrinsic and acquired antibiotic resistance (Carmeli et al. 1999; Lister, Wolter, and Hanson 2009; Chambers and Deleo 2009; Lowy 2003), making infections by these pathogens increasingly difficult to treat. Adding to the complexity of managing these infections is the newly acknowledged phenomenon that co-cultivation of these species enhances survival in the presence of antimicrobials. Synergistic effects of co-culture are observed when assessing tolerance of S. aureus to both gentamicin and tetracycline when grown either in planktonic culture or in an in vitro wound model (DeLeon et al. 2014). Hoffman, et al, further demonstrated that increased tolerance of S. aureus to tobramycin during co-culture occurs through the secretion of P. aeruginosa exoproducts called alkyl-quinolones (AQs) (Hoffman et al. 2006), secondary metabolites that mediate interactions of this pathogen with several other microbial species (Nguyen et al. 2015; Filkins et al. 2015; Mashburn et al. 2005; Korgaonkar and Whiteley 2011; Heeb et al. 2011) (Table 1).
Table 1.
Structure and functions of key alkyl-quinolones
| Nomenclature | Functions | Structure | |
|---|---|---|---|
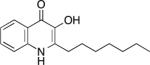
| PQS | 2- heptyl-3-hydroxy-4(1H)-quinolone (Pseudomonas quinolone signal) | • quorum sensing - virulence gene expression • iron chelation • vesicle formation |
|
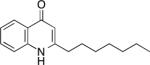
| HHQ | 2-heptyl-4-hydroxyquinoline | • quorum sensing - virulence gene expression • signaling - phenazine production |
|
| HQNO | 2-heptyl-4-hydroxyquinoline N-oxide | • cytochrome bc1 complex inhibitor |
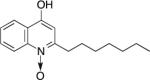
|
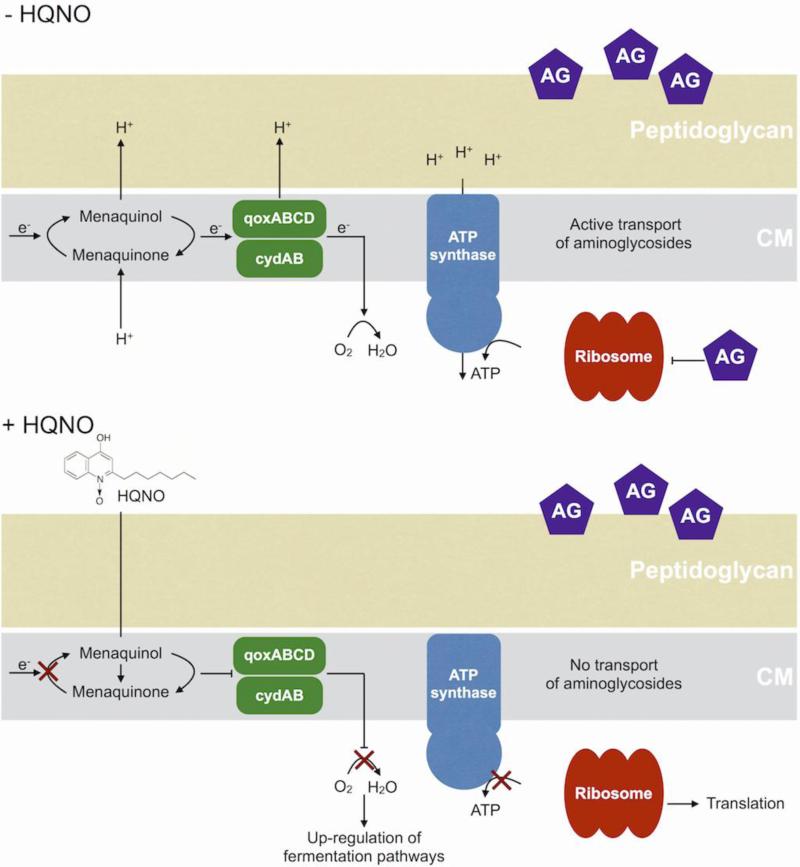
One specific AQ, 4-hydroxy-2-heptylquinoline-N-oxide (HQNO), acts as a quinone analog, binding to and inhibiting activity of cytochrome b in Gram-positive bacteria such as S. aureus (Machan et al. 1992; Lightbown and Jackson 1956) (Fig. 1). Inhibition of cytochrome b results in decreased electron transport and thus energy production (Lightbown and Jackson 1956; Gotz and Mayer 2013). This reduction in ATP production shifts S. aureus metabolism from aerobic respiration to anaerobic or fermentative respiration (Hoffman et al. 2006; Filkins et al. 2015; Biswas et al. 2009). By inhibiting respiration, long term exposure to HQNO selects for S. aureus small colony variants (SCVs), producing a similar outcome as either mutations in the terminal oxidases QoxABCD and CydAB (Hammer et al. 2013) or exposure to secondary metabolites such as pyocyanin (Voggu et al. 2006; Biswas et al. 2009). SCV formation in S. aureus also increases tolerance to aminoglycoside antibiotics, which require an active electron transport chain to enter the cell (Bryan and Van Den Elzen 1977) (Fig. 1). Therefore, SCVs have been shown to display increased tolerance to multiple antimicrobials (Pan et al. 2002; Proctor et al. 2014; Proctor et al. 2006; Lechner, Lewis, and Bertram 2012; Wood, Knabel, and Kwan 2013).
Figure 1. HQNO inhibits cytochrome b and increases tolerance to antimicrobials.
Figure showing cytoplasmic membrane (CM) of S. aureus. HQNO is a menaquinone analog that inhibits electron transport through cytochrome b. This results in decreased ATP generation and a shift to fermentative metabolism. Reduction of ATP in the cell decreases active transport which is required for uptake of aminoglycoside (AG) antibiotics.
Co-culture with S. aureus similarly promotes selection of P. aeruginosa SCVs, resulting in increased survival and antimicrobial tolerance of P. aeruginosa isolates in the CF lung (Michelsen et al. 2014). This interaction is dependent upon the Agr quorum sensing system, which regulates the expression of multiple virulence factors in S. aureus (Novick 2003). The precise mechanism by which S. aureus quorum sensing affects P. aeruginosa antimicrobial tolerance however remains unclear.
Another major contributor to antibiotic tolerance and virulence during polymicrobial infections is biofilm formation. Mixed microbial biofilms are especially relevant in the context of CF due to the altered physiology of the lung (Costerton 2001; Costerton et al. 1995). Thick, dehydrated mucus and hypoxia in the CF lung provide prime conditions for biofilm formation by both S. aureus and P. aeruginosa (Costerton 2001). A recent report by Fugere et al., showed that production of HQNO and the pseudomonas quinolone signal (PQS), another AQ produced by P. aeruginosa, enhances biofilm formation by S. aureus (Fugere et al. 2014). The promotion of mixed biofilms by these species results in decreased susceptibility to multiple classes of antibiotics, complicating eradication of co-colonizing microorganisms.
PARASITIC INTERACTIONS
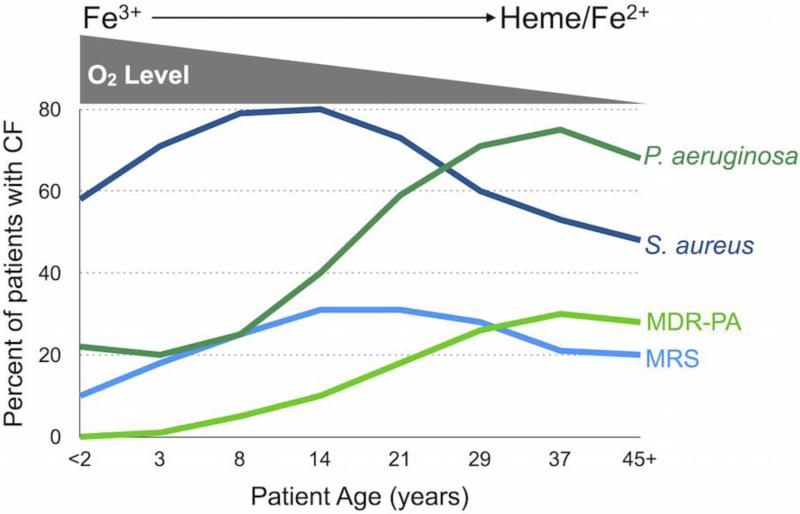
In 1949, Jacques Monod proposed that limited availability of required nutrients shapes the populations of competing microbes (Monod 1949). Such a change in population dynamics is often observed during chronic CF lung infections. The microbial makeup of the lungs is highly diverse and dynamic in young CF patients, yet this environment becomes dominated by P. aeruginosa as disease progresses (Foundation 2014) (Fig. 2). Many factors impact the makeup of the bacterial population that inhabit this environment, including changes in nutrient availability and the host immune system (Quinn et al. 2014). The studies highlighted below outline how specific host factors drive microbial interactions during polymicrobial infections.
Figure 2. P. aeruginosa displaces S. aureus in the CF lung.
P. aeruginosa and multi-drug resistant P. aeruginosa (MDR-PA) outcompete S. aureus and methicillin-resistant S. aureus (MRSA) over the course of CF lung infection. As CF lung disease progresses, oxygen levels decrease coinciding with the use of heme and ferrous iron as predominant sources of iron.
Iron availability
Iron is an essential nutrient for most microbial pathogens and is increasingly appreciated as a critical mediator of CF lung disease (Bouvier 2016; Barnabie and Whiteley 2015; Cassat and Skaar 2013). Iron is limiting during infections due to sequestration by host proteins such as lactoferrin, transferrin, and hemoglobin (Otto, Verweij-van Vught, and MacLaren 1992; Nairz et al. 2010). Microbial pathogens have therefore evolved several means of acquiring this nutrient from the host. P. aeruginosa and S. aureus obtain iron through the secretion of siderophores (Ochsner, Vasil, and Vasil 1995; Martin et al. 2011; Hammer and Skaar 2011), heme transport and degradation (Ochsner, Johnson, and Vasil 2000; Maresso and Schneewind 2006), and ferrous iron uptake systems (Wang et al. 2011; Ster et al. 2010). While siderophore-mediated iron uptake is clearly important for acute P. aeruginosa infections (Takase et al. 2000; Meyer 2000; Meyer et al. 1997), several recent reports demonstrate a decreased reliance on siderophores and an increased utilization of heme and ferrous as an iron source by P. aeruginosa during CF infection (Nguyen et al. 2014; Huse et al. 2010; Konings et al. 2013; Hunter et al. 2013) (Fig. 2). Skaar et al. further showed that S. aureus preferentially imports heme over siderophore-mediated scavenging of iron from transferrin (Skaar et al. 2004). Thus, P. aeruginosa and S. aureus likely compete for limiting and overlapping sources of iron during polymicrobial infections.
While the majority of microbial iron uptake studies have been performed with pure cultures, recent studies are yielding new insights into how co-culture impacts iron homeostasis. P. aeruginosa was previously shown to lyse S. aureus, thus liberating iron from this competing microbial species during co-culture (Mashburn et al. 2005). This activity requires the P. aeruginosa pqsA gene, encoding the first enzyme in AQ biosynthesis, demonstrating a role for AQs in this parasitic interaction. More recently, our lab showed that iron deprivation enhances the production of certain AQs, correlating with increased antagonism of S. aureus growth in low iron environments (Nguyen et al. 2015). Filkins et al., also showed that HQNO produced by P. aeruginosa drives S. aureus towards fermentative metabolism when the two species are grown in mixed biofilms, resulting in eventual reduced viability of S. aureus (Filkins et al. 2015). The authors additionally showed that this parasitic behavior requires siderophore-mediated iron uptake by P. aeruginosa, again demonstrating a role for iron in this parasitic relationship. Release of peptidoglycan upon S. aureus lysis during co-culture further enhances AQ production by P. aeruginosa (Korgaonkar and Whiteley 2011), presumably allowing for a positive feedback of each of these parasitic behaviors. These studies collectively demonstrate the potential for nutrient availability, as well as microbemicrobe sensing, to enhance parasitic behaviors during polymicrobial infections.
Host immunity
In order to survive during polymicrobial infections, competing microbes must also be able to circumvent the host immune system. Recent studies suggest modulation of the immune system by co-infecting pathogens affects the make up of polymicrobial infections. While the above studies show that changes in nutrient availability, particularly iron, likely contribute to this shift, modulation of host immunity likely also plays a role. In line with this idea, P. aeruginosa induces the production of type-IIA-secreted PLA2 (sPLA2-IIA), a potent bactericidal enzyme, by CF lung cells (Pernet et al. 2014). Notably, the levels detected in CF sputum are potent enough to kill S. aureus but have minimal effects on P. aeruginosa viability (Pernet et al. 2014). Thus, P. aeruginosa appears to induce host bacteriostatic factors to levels that specifically inhibit the growth of competing microbes.
P. aeruginosa is also capable of degrading host immune factors, including those involved in the sequestration of iron. Lactoferrin binds to the oxidized, insoluble form of ferric iron (Fe3+) at mucosal surfaces, sequestering this nutrient from infecting microbes (Nairz et al. 2010). The high affinity of secreted siderophores for Fe3+ allows this nutrient to be scavenged by microbial pathogens, overcoming this barrier to infection (Otto, Verweij-van Vught, and MacLaren 1992). Lactoferrin is degraded in CF lungs colonized by P. aeruginosa, which promotes biofilm formation by this pathogen (Rogan et al. 2004). Biofilm formation in turn contributes to increased local hypoxia of the CF lung, correlating with increased dependency on systems that mediate the uptake of reduced, ferrous iron (Fe2+) by P. aeruginosa (Hunter et al. 2013). The impact of decreased lactoferrin on the survival of other microbial species in the CF lung has not yet been investigated. However, one potential hypothesis is that degradation of lactoferrin combined with increased hypoxia of the CF lung confer a specific advantage to P. aeruginosa over CF pathogens that are not as adept at ferrous iron uptake.
Chronic polymicrobial infections involving P. aeruginosa and S. aureus are often the result of a dysfunctional or depressed immune system, and the long-term nature of these infections further alters host physiology and immunity (Peters et al. 2012). Analysis of the DK-2 lineage of P. aeruginosa, isolated from multiple CF patients between 1973 and 2008 (Yang et al. 2011), showed that this strain's ability to inhibit S. aureus growth was reduced as it adapted to the CF lung environment (Michelsen et al. 2014). A separate analysis of 24 P. aeruginosa isolates from 8 distinct CF patients at multiple stages of chronic lung infection showed a similar reduction in the ability of P. aeruginosa to inhibit S. aureus growth (Baldan et al. 2014). Thus, adaptation to the host environment during chronic CF lung infections shifts the relationship between these two species from parasitic to commensal.
OUTCOMES OF S. AUREUS-P. AERUGINOSA INTERACTIONS
The above highlighted microbial interactions clearly have the potential to exert synergistic impacts on the progression of polymicrobial infections. The most obvious of these is how increased antimicrobial tolerance during co-cultivation can complicate therapeutic management of co-infections. Enhanced virulence also occurs through the increased production of AQ metabolites that mediate both parasitic and mutualistic interactions. PQS functions as a quorum sensing molecule to activate the production of lytic enzymes that promote tissue invasion and redox-active phenazines that exhibit toxicity against eukaryotic cells (Pesci et al. 1999; Gallagher et al. 2002) (Table 1). The precursor to PQS, HHQ, also induces the expression of lytic enzymes and phenazines (Diggle et al. 2007; Deziel et al. 2004) (Table 1). As such, enhanced synthesis and secretion of AQs in response to other microbial pathogens promotes increased production of virulence factors by P. aeruginosa (Korgaonkar et al. 2013).
Several infection models have been developed to investigate the impact of co-infections of P. aeruginosa and S. aureus, including the impact of AQs on virulence. A Drosophila model was used to screen 40 oropharngeal isolates, including some Staphylococcal species, for virulence in the presence and absence of P. aeruginosa, resulting in the identification of three distinct infection classes (Sibley et al. 2008). Class 3 represented isolates that were avirulent in mono-culture, but when co-cultured with P. aeruginosa became pathogenic. This model system was also used to show that P. aeruginosa increased AQ and virulence factor production in the presence of peptidoglycan released from S. aureus, resulting in increased mortality of the fly (Korgaonkar et al. 2013; Korgaonkar and Whiteley 2011). A mammalian polymicrobial infection model, in which mixed in vitro biofilms are transplanted into the wounds of mice, has also been used to study co-infections of S. aureus and P. aeruginosa (Dalton et al. 2011). The authors of this study found that wound closure was delayed, and antimicrobial therapy was less effective, in S. aureus-P. aeruginosa co-infections as compared to mono-infection with P. aeruginosa. Combined, these studies highlight the potential for increased virulence due to co-infections.
While these studies demonstrate that co-infection often results in enhanced virulence, variations in host age, overall health status, and other microbial inhabitants complicate the ability to predict the outcomes of polymicrobial infections. This is particularly true in the case of CF lung infections, in which both synergistic and antagonistic outcomes of co-infection have been noted. According to a predictive model by Liou et al., the presence of S. aureus increases survivorship of patients co-infected with P. aeruginosa, suggesting interactions between these two pathogens have beneficial consequences for the infected host (Liou et al. 2001). However, other reports show that the presence of methicillin resistant S. aureus (MRSA) with P. aeruginosa correlates with increased morbidity and mortality for co-infected CF patients due to increased antibiotic resistance and rate of lung function decline (Hubert et al. 2013). A recent study examined S. aureus as a marker for CF lung disease in adult patients. In children with CF disease, the presence of S. aureus correlated with increased inflammation and decreased lung function (Ahlgren et al. 2015). In contrast, S. aureus in adults could be a marker for milder disease, particularly if it coincides with reduced colonization by P. aeruginosa. Thus, host-specific factors play a critical role in the manifestation of polymicrobial infections involving S. aureus and P. aeruginosa.
Even when analyzing co-cultures in vitro, defining whether specific interactions between S. aureus and P. aeruginosa are parasitic or mutualistic is not as simple or clear as it may first appear. As discussed earlier in this review, P. aeruginosa produces and secretes a secondary metabolite, HQNO, that suppresses S. aureus growth by inhibiting respiration. While this behavior is seemingly parasitic, as growth suppression would allow P. aeruginosa to outcompete S. aureus for essential nutrients, it alsoselects for small colony variants of S. aureus, increasing the potential for this competing microbe to persist during infection (Hoffman et al. 2006). Studies cited above from Michelsen, et al., further show that the reduced ability of P. aeruginosa CF isolates to inhibit S. aureus growth correlates with the selection of P. aeruginosa isolates with increased antibiotic tolerance during co-culture (Michelsen et al. 2014). More studies into the long-term outcomes of chronic polymicrobial infections, relying on appropriate in vitro and in vivo models, are clearly needed to improve our understanding of how these interactions contribute to infectious disease.
CONCLUSION
Regardless of how interactions between S. aureus and P. aeruginosa are characterized, the presence of these pathogens together clearly results in distinct disease manifestations and warrants careful consideration for successful management of polymicrobial infections. As highlighted in this review, co-culture of S. aureus with P. aeruginosa affects the virulence capacity and vulnerability to antimicrobial therapy of both pathogens, exerting both synergistic and antagonistic effects on disease progression. Future studies defining the impacts of interspecies interactions, as well as the host factors that modulate these interactions, are critical for understanding the pathogenesis of polymicrobial infections.
Acknowledgments
Funding is provided by the University of Maryland School of Pharmacy (to AGO) and NIH training grant T32 GM 066706 (to ATN).
Footnotes
COMPLIANCE WITH ETHICAL STANDARDS
Angela T. Nguyen declares that she has no conflict of interest. Amanda G. Oglesby-Sherrouse declares that she has no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors.
REFERENCES
- Ahlgren HG, Benedetti A, Landry JS, Bernier J, Matouk E, Radzioch D, Lands LC, Rousseau S, Nguyen D. Clinical outcomes associated with Staphylococcus aureus and Pseudomonas aeruginosa airway infections in adult cystic fibrosis patients. BMC Pulm Med. 2015;15:67. doi: 10.1186/s12890-015-0062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldan R, Cigana C, Testa F, Bianconi I, De Simone M, Pellin D, Di Serio C, Bragonzi A, Cirillo DM. Adaptation of Pseudomonas aeruginosa in Cystic Fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One. 2014;9:e89614. doi: 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnabie PM, Whiteley M. Iron-Mediated Control of Pseudomonas aeruginosa-Staphylococcus aureus Interactions in the Cystic Fibrosis Lung. J Bacteriol. 2015;197:2250–1. doi: 10.1128/JB.00303-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas L, Biswas R, Schlag M, Bertram R, Gotz F. Small-colony variant selection as a survival strategy for Staphylococcus aureus in the presence of Pseudomonas aeruginosa. Appl Environ Microbiol. 2009;75:6910–2. doi: 10.1128/AEM.01211-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier NM. Cystic fibrosis and the war for iron at the host-pathogen battlefront. Proc Natl Acad Sci U S A. 2016 doi: 10.1073/pnas.1525101113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan LE, Van Den Elzen HM. Effects of membrane-energy mutations and cations on streptomycin and gentamicin accumulation by bacteria: a model for entry of streptomycin and gentamicin in susceptible and resistant bacteria. Antimicrob Agents Chemother. 1977;12:163–77. doi: 10.1128/aac.12.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeli Y, Troillet N, Eliopoulos GM, Samore MH. Emergence of antibiotic-resistant Pseudomonas aeruginosa: comparison of risks associated with different antipseudomonal agents. Antimicrob Agents Chemother. 1999;43:1379–82. doi: 10.1128/aac.43.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–19. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9:50–2. doi: 10.1016/s0966-842x(00)01918-1. [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–45. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One. 2011;6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun. 2014;82:4718–28. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E, Lepine F, Milot S, He J, Mindrinos MN, Tompkins RG, Rahme LG. Analysis of Pseudomonas aeruginosa 4-hydroxy-2-alkylquinolines (HAQs) reveals a role for 4-hydroxy-2-heptylquinoline in cell-to-cell communication. Proc Natl Acad Sci U S A. 2004;101:1339–44. doi: 10.1073/pnas.0307694100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle SP, Matthijs S, Wright VJ, Fletcher MP, Chhabra SR, Lamont IL, Kong X, Hider RC, Cornelis P, Camara M, Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Filkins LM, Graber JA, Olson DG, Dolben EL, Lynd LR, Bhuju S, O'Toole GA. Co-culture of Staphylococcus aureus with Pseudomonas aeruginosa drives S. aureus towards fermentative metabolism and reduced viability in a cystic fibrosis model. J Bacteriol. 2015 doi: 10.1128/JB.00059-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filkins LM, O'Toole GA. Cystic Fibrosis Lung Infections: Polymicrobial, Complex, and Hard to Treat. PLoS Pathog. 2015;11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foundation, Cystic Fibrosis . Patient Registry Annual Data Report 2014. Cystic Fibrosis Foundation; Bethesda, Maryland: 2014. [Google Scholar]
- Fugere A, Lalonde Seguin D, Mitchell G, Deziel E, Dekimpe V, Cantin AM, Frost E, Malouin F. Interspecific small molecule interactions between clinical isolates of Pseudomonas aeruginosa and Staphylococcus aureus from adult cystic fibrosis patients. PLoS One. 2014;9:e86705. doi: 10.1371/journal.pone.0086705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher LA, McKnight SL, Kuznetsova MS, Pesci EC, Manoil C. Functions required for extracellular quinolone signaling by Pseudomonas aeruginosa. J Bacteriol. 2002;184:6472–80. doi: 10.1128/JB.184.23.6472-6480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz F, Mayer S. Both terminal oxidases contribute to fitness and virulence during organ-specific Staphylococcus aureus colonization. Mbio. 2013;4:e00976–13. doi: 10.1128/mBio.00976-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Reniere ML, Cassat JE, Zhang Y, Hirsch AO, Indriati Hood M, Skaar EP. Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. Mbio. 2013;4 doi: 10.1128/mBio.00241-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65:129–47. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Camara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol Rev. 2011;35:247–74. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, Deziel E, D'Argenio DA, Lepine F, Emerson J, McNamara S, Gibson RL, Ramsey BW, Miller SI. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103:19890–5. doi: 10.1073/pnas.0606756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert D, Reglier-Poupet H, Sermet-Gaudelus I, Ferroni A, Le Bourgeois M, Burgel PR, Serreau R, Dusser D, Poyart C, Coste J. Association between Staphylococcus aureus alone or combined with Pseudomonas aeruginosa and the clinical condition of patients with cystic fibrosis. Journal of Cystic Fibrosis. 2013;12:497–503. doi: 10.1016/j.jcf.2012.12.003. [DOI] [PubMed] [Google Scholar]
- Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot A, Cornelis P, Newman DK. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. Mbio. 2013;4 doi: 10.1128/mBio.00557-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse HK, Kwon T, Zlosnik JEA, Speert DP, Marcotte EM, Whiteley M. Parallel Evolution in Pseudomonas aeruginosa over 39,000 Generations In Vivo. Mbio. 2010;1 doi: 10.1128/mBio.00199-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings AF, Martin LW, Sharples KJ, Roddam LF, Latham R, Reid DW, Lamont IL. Pseudomonas aeruginosa uses multiple pathways to acquire iron during chronic infection in cystic fibrosis lungs. Infect Immun. 2013;81:2697–704. doi: 10.1128/IAI.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar AK, Whiteley M. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol. 2011;193:909–17. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A. 2013;110:1059–64. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner S, Lewis K, Bertram R. Staphylococcus aureus persisters tolerant to bactericidal antibiotics. J Mol Microbiol Biotechnol. 2012;22:235–44. doi: 10.1159/000342449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightbown JW, Jackson FL. Inhibition of cytochrome systems of heart muscle and certain bacteria by the antagonists of dihydrostreptomycin: 2-alkyl-4-hydroxyquinoline N-oxides. Biochem J. 1956;63:130–7. doi: 10.1042/bj0630130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou TG, Adler FR, Fitzsimmons SC, Cahill BC, Hibbs JR, Marshall BC. Predictive 5-year survivorship model of cystic fibrosis. Am J Epidemiol. 2001;153:345–52. doi: 10.1093/aje/153.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22:582–610. doi: 10.1128/CMR.00040-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111:1265–73. doi: 10.1172/JCI18535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SV, Bruce KD. The cystic fibrosis airway microbiome. Cold Spring Harb Perspect Med. 2013;3:a009738. doi: 10.1101/cshperspect.a009738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machan ZA, Taylor GW, Pitt TL, Cole PJ, Wilson R. 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1992;30:615–23. doi: 10.1093/jac/30.5.615. [DOI] [PubMed] [Google Scholar]
- Maresso AW, Schneewind O. Iron acquisition and transport in Staphylococcus aureus. Biometals. 2006;19:193–203. doi: 10.1007/s10534-005-4863-7. [DOI] [PubMed] [Google Scholar]
- Marsland BJ, Gollwitzer ES. Host-microorganism interactions in lung diseases. Nat Rev Immunol. 2014;14:827–35. doi: 10.1038/nri3769. [DOI] [PubMed] [Google Scholar]
- Martin LW, Reid DW, Sharples KJ, Lamont IL. Pseudomonas siderophores in the sputum of patients with cystic fibrosis. Biometals. 2011;24:1059–67. doi: 10.1007/s10534-011-9464-z. [DOI] [PubMed] [Google Scholar]
- Mashburn LM, Jett AM, Akins DR, Whiteley M. Staphylococcus aureus serves as an iron source for Pseudomonas aeruginosa during in vivo coculture. J Bacteriol. 2005;187:554–66. doi: 10.1128/JB.187.2.554-566.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM. Pyoverdines: pigments, siderophores and potential taxonomic markers of fluorescent Pseudomonas species. Arch Microbiol. 2000;174:135–42. doi: 10.1007/s002030000188. [DOI] [PubMed] [Google Scholar]
- Meyer JM, Stintzi A, De Vos D, Cornelis P, Tappe R, Taraz K, Budzikiewicz H. Use of siderophores to type pseudomonads: the three Pseudomonas aeruginosa pyoverdine systems. Microbiology. 1997;143(Pt 1):35–43. doi: 10.1099/00221287-143-1-35. [DOI] [PubMed] [Google Scholar]
- Michelsen CF, Christensen AM, Bojer MS, Hoiby N, Ingmer H, Jelsbak L. Staphylococcus aureus alters growth activity, autolysis, and antibiotic tolerance in a human host-adapted Pseudomonas aeruginosa lineage. J Bacteriol. 2014;196:3903–11. doi: 10.1128/JB.02006-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monod J. The Growth of Bacterial Cultures. Annual Review of Microbiology. 1949;3:371–94. [Google Scholar]
- Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52:188–99. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nairz M, Schroll A, Sonnweber T, Weiss G. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol. 2010;12:1691–702. doi: 10.1111/j.1462-5822.2010.01529.x. [DOI] [PubMed] [Google Scholar]
- Nguyen AT, Jones JW, Ruge MA, Kane MA, Oglesby-Sherrouse AG. Iron depletion enhances production of antimicrobials by Pseudomonas aeruginosa. J Bacteriol. 2015 doi: 10.1128/JB.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, O'Neill MJ, Watts AM, Robson CL, Lamont IL, Wilks A, Oglesby-Sherrouse AG. Adaptation of iron homeostasis pathways by a Pseudomonas aeruginosa pyoverdine mutant in the cystic fibrosis lung. J Bacteriol. 2014 doi: 10.1128/JB.01491-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–49. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Johnson Z, Vasil ML. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146(Pt 1):185–98. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- Ochsner UA, Vasil AI, Vasil ML. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: purification and activity on iron-regulated promoters. J Bacteriol. 1995;177:7194–201. doi: 10.1128/jb.177.24.7194-7201.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto BR, Verweij-van Vught AM, MacLaren DM. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–33. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- Pan XS, Hamlyn PJ, Talens-Visconti R, Alovero FL, Manzo RH, Fisher LM. Small-colony mutants of Staphylococcus aureus allow selection of gyrase-mediated resistance to dual-target fluoroquinolones. Antimicrob Agents Chemother. 2002;46:2498–506. doi: 10.1128/AAC.46.8.2498-2506.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet E, Guillemot L, Burgel PR, Martin C, Lambeau G, Sermet-Gaudelus I, Sands D, Leduc D, Morand PC, Jeammet L, Chignard M, Wu Y, Touqui L. Pseudomonas aeruginosa eradicates Staphylococcus aureus by manipulating the host immunity. Nat Commun. 2014;5:5105. doi: 10.1038/ncomms6105. [DOI] [PubMed] [Google Scholar]
- Pesci EC, Milbank JB, Pearson JP, McKnight S, Kende AS, Greenberg EP, Iglewski BH. Quinolone signaling in the cell-to-cell communication system of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1999;96:11229–34. doi: 10.1073/pnas.96.20.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters BM, Jabra-Rizk MA, O'May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price KE, Hampton TH, Gifford AH, Dolben EL, Hogan DA, Morrison HG, Sogin ML, O'Toole GA. Unique microbial communities persist in individual cystic fibrosis patients throughout a clinical exacerbation. Microbiome. 2013;1:27. doi: 10.1186/2049-2618-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RA, Kriegeskorte A, Kahl BC, Becker K, Loffler B, Peters G. Staphylococcus aureus Small Colony Variants (SCVs): a road map for the metabolic pathways involved in persistent infections. Front Cell Infect Microbiol. 2014;4:99. doi: 10.3389/fcimb.2014.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- Quinn RA, Lim YW, Maughan H, Conrad D, Rohwer F, Whiteson KL. Biogeochemical forces shape the composition and physiology of polymicrobial communities in the cystic fibrosis lung. Mbio. 2014;5:e00956–13. doi: 10.1128/mBio.00956-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan MP, Taggart CC, Greene CM, Murphy PG, O'Neill SJ, McElvaney NG. Loss of microbicidal activity and increased formation of biofilm due to decreased lactoferrin activity in patients with cystic fibrosis. J Infect Dis. 2004;190:1245–53. doi: 10.1086/423821. [DOI] [PubMed] [Google Scholar]
- Rogers GB, Hoffman LR, Whiteley M, Daniels TW, Carroll MP, Bruce KD. Revealing the dynamics of polymicrobial infections: implications for antibiotic therapy. Trends Microbiol. 2010;18:357–64. doi: 10.1016/j.tim.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–8. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ster C, Beaudoin F, Diarra MS, Jacques M, Malouin F, Lacasse P. Evaluation of some Staphylococcus aureus iron-regulated proteins as vaccine targets. Vet Immunol Immunopathol. 2010;136:311–8. doi: 10.1016/j.vetimm.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Surette MG. The Cystic Fibrosis Lung Microbiome. American Thoracic Society. 2013;11:61–65. doi: 10.1513/AnnalsATS.201306-159MG. [DOI] [PubMed] [Google Scholar]
- Takase H, Nitanai H, Hoshino K, Otani T. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect Immun. 2000;68:1834–9. doi: 10.1128/iai.68.4.1834-1839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, Gotz F. Microevolution of cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol. 2006;188:8079–86. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol. 2011;193:3606–17. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood TK, Knabel SJ, Kwan BW. Bacterial persister cell formation and dormancy. Appl Environ Microbiol. 2013;79:7116–21. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Jelsbak L, Marvig RL, Damkiaer S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Hoiby N, Sommer MO, Molin S. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A. 2011;108:7481–6. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]