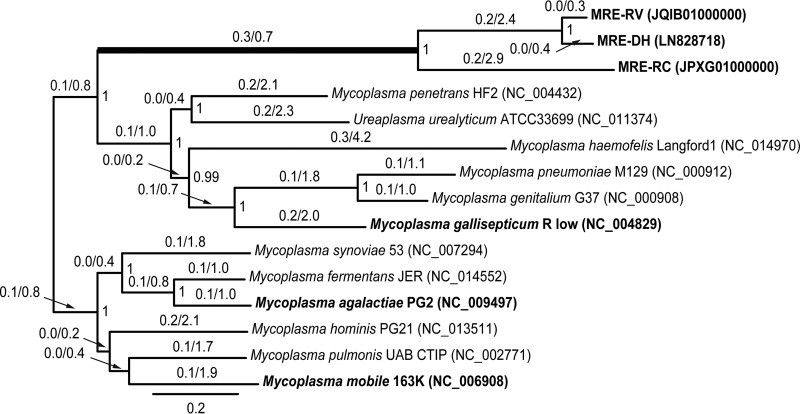
FIG 1 .
Bayesian phylogeny of MRE reconstructed based on the concatenated nucleotide sequences of the following genes: dnaG, infC, nusA, rplA, rplB, rplC, rplE, rplF, rplM, rplN, rplP, rplT, rpmA, rpsB, rpsC, rpsE, rpsJ, rpsS, and smpB. MRE were sampled from Dentiscutata heterogama (MRE-DH), Racocetra verrucosa (MRE-RV), and Rhizophagus clarus (MRE-RC). Bayesian posterior probabilities greater than 0.90 are shown at nodes; Bayesian analyses were conducted using MrBayes v.3.2 (64) under the nucleotide substitution model GTR + Γ + I with 1,000,000 generations and a 250,000 burn-in. The numbers above branches are the estimates of dN/dS ratios obtained using the codeml module of PAML v.4.8 (65), assuming a two-ratio model. The thickened branch leading to MRE indicates that, according to the likelihood ratio model testing conducted in codeml, the dN/dS ratio along it is significantly different from the background dN/dS ratio for all other branches (χ2 = 36.12, P < 0.001). Taxa in boldface were subjected to Tajima 1D relative rate tests (26) (Table 1).