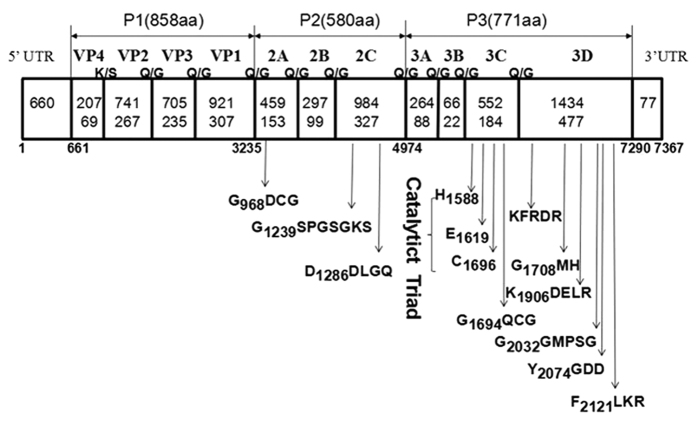
Figure 1. Genomic organisation of SEV-gx with its predicted cleavage sites and conserved Enterovirus motifs.
The open reading frame is flanked by the 5′-untranslated region (UTR) and the 3′-UTR. Capsid proteins of P1 (VP4, VP2, VP3 and VP1) and nonstructural proteins of P2 (2A, 2B and 2C) and P3 (3A, 3B, 3C and 3D) are shown with nucleotide lengths (upper) and amino acid lengths (lower). The positions of some motifs are shown by the first position in the motif.