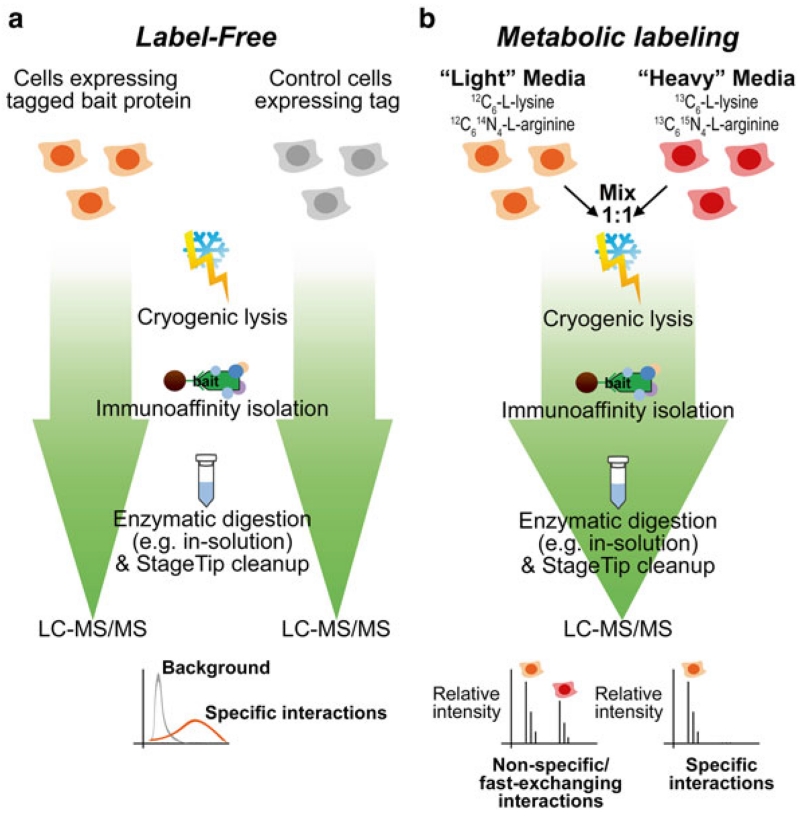
Fig. 1.
Integrated label-free and metabolic label-based approach for profiling protein interactions. (a) Label-free workflow. Cells expressing the tagged protein of interest (bait protein) are cultured in parallel to cells expressing the tag alone. Following cryogenic lysis, immunoaffinity isolation of the tagged protein (with its interactions) is performed using antibody-conjugated magnetic beads. Captured proteins are subjected to enzymatic digestion, sample clean-up and mass spectrometry analysis. Mass spectrometry signals (e.g., spectrum counts or peak intensities) from the bait protein isolation versus control isolations are analyzed to set cut-offs for high-confidence specific interactions. (b) Metabolic labeling workflow. Cells expressing tagged bait protein are cultured in media containing “light” amino acids, while control wild type cells (not expressing a tag) are cultured in “heavy” amino acid media. Cells are mixed in a 1:1 ratio and subjected to cryogenic lysis, immunoaffinity purification, and proteomic analysis as in (a), except the MS data analysis calculates the relative ion intensities of the “heavy” and “light” peptide signals to determine the specificity of interactions