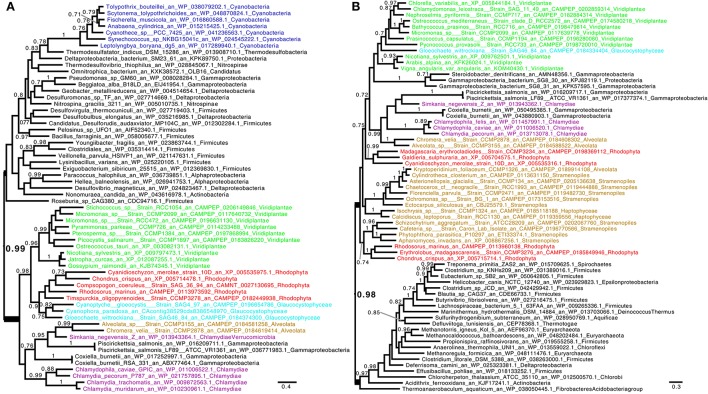
Figure 1.
Consensus tree obtained with Phylobayes 4.1 with Bayesian posterior probabilities mapped onto the nodes of (A) phosphoribosylanthranilate isomerase (TrpF) and (B) anthranilate phosphoribosyltransferase (TrpD). Groups of interest are highlighted in purple (Chlamydiales), green (green algae and plants), red (red algae), cyan (Glaucophyta), and blue (Cyanobacteria). Lineages putatively derived from secondary endosymbiosis of Archaeplastida are displayed in brown. Bayesian posterior probabilities values (PP) higher than 0.7 are indicated onto the branches. The scale bars indicate the inferred number of amino acid substitutions per site. The trees were manually rooted for convenience of display. The nodes uniting Archaeplastida and their derived lineages to Chlamydiales by LGT are highlighted in bold. The Phosphoribosylanthranilate isomerase tree (A) shows a group with robust support (PP = 0.99) composed of Chlamydiales, all Archaeplastida, Alveolata, and the two intracellular γ-proteobacteria pathogens Coxiella and Piscirickettsia. Bonner et al. (2014) have proposed that Coxiella received its Tryptophan operon through LGT from Simkaniaceae (Chlamydiales) in a common intracellular environment (see text). (B) shows that the three Archaeplastida lineages are united through LGT together with a complex pattern of secondary endosymbiosis lineages (PP = 0.98). Here again intracellular γ-proteobacteria are recovered for the same reasons. Sequences used in the trees were retrieved by homology searches with BLAST against sequences of interest in a database composed of nr, MMETSP (Keeling et al., 2014), and genomes of interest. Sequences with an E < 1e-10 were selected and aligned using MAFFT (Katoh and Standley, 2013). We used BMGE (Criscuolo and Gribaldo, 2010) with a block size of four and the BLOSUM30 similarity matrix for block selection. We generated preliminary trees using Fasttree (Price et al., 2010). “Dereplication,” using TreeTrimmer (Maruyama et al., 2013), was applied to supported monophyletic clades in order to reduce sequence redundancy. The final set of sequences were selected manually. Finally, the sequences were re-aligned with MUSCLE (Edgar, 2004), block selection was carried out using BMGE with the same setting, and trees were generated using Phylobayes 4.1 (Lartillot et al., 2009) under the CAT+GTR model (Lartillot and Philippe, 2004). The two chains were stopped when convergence was reached (maxdiff < 0.1) after at least 300 cycles and a burn-in different for each alignment when equilibrium was reached between two chains.