Summary
The recognition of CD1-lipid complexes by T cells was discovered twenty years ago and has since been an emerging and expanding field of investigation. Unlike protein antigens, which are presented on MHC class I and II molecules, lipids can only be presented by CD1 molecules, a unique family of MHC-like proteins whose singularity is a hydrophobic antigen binding groove. The processing and loading of lipid antigens inside this groove of CD1 molecules require localization to late endosomal and lysosomal subcellular compartments and their acidic pHs. This particular environment provides the necessary glycolytic enzymes and lipases that process lipid and glycolipid antigens, as well as a set of lipid transfer proteins that load the final version of the antigen inside the groove of CD1. The overall sequence of events needed for efficient presentation of lipid antigens is now understood and presented in this review. However, a large number of important details have been elusive. This elusiveness is linked to the inherent technical difficulties of studying lipids and the lipid-protein interface in vitro and in vivo. Here, we will expose some of those limitations and describe new approaches to address them during the characterization of lipids and glycolipids antigen presentation.
Keywords: Natural Killer T cells, Lipid Mediators, T Cell Receptors, Antigen Presentation/Processing, CD1
Introduction
Since its inception in the 1980s, the field of antigen presentation has been dominated by studies that have examined processing and loading of peptide fragments onto major histocompatibility complex (MHC) molecules for presentation to T cells. No or little interest was granted to glycans as antigenic structures because of the T-cell independent nature of the immune recognition towards them, whilst lipids in general were considered only as accidental antigenic structures. The discovery of CD1, a β2-microglobulin-associated MHC Class I-like molecule in 1986, which as we will expose below in this review is central to lipid presentation, did not change this situation for ten years. In hindsight, it is striking to remember that antigen presentation was so obsessed with peptides that the first antigen isolated for CD1d was a peptide(1). It took two more years and the combination in 1994–1995 of seminal structural work by the Wilson’s group(2) and clever biochemistry and cellular immunology by the Brenner’s group(3) to come to the conclusion that indeed CD1 molecules were presenting lipid antigens to T cells. The interest in the processing and presentation of lipid structures to the immune system has steadily grown over the past twenty years using two principal models to understand the functional consequences of presentation by CD1 to T cells: NKT cells(4) and mycobacterial infections(5). However, progress has been slowed by technical limitations that come from two fronts: the lack of sensitivity of lipid analytical methods and the difficulty of the organic synthetic chemistry of lipids, especially glycolipids. These persistent issues have resulted in difficulty in determining the specificity of anti-lipid T cells and following their fate. We will highlight in this review the classical views of lipid antigen processing and presentation as well as areas where new approaches and new reagents should allow simpler determination of glycolipid chemical structures to understand CD1 biology.
The Discovery of CD1 Genes and Products
The human CD1 gene cluster was first discovered in 1986 and was described as being related to MHC despite being located on a different chromosome (6). Although initially considered to be the human version of the mouse Thymus Leukemia (TL) antigens, CD1 genes and products were later found in the murine thymus (7) and rapidly organized based on sequence homologies into two classes (8): group 1, inclusive of CD1a, b, c, and e, and group 2, composed of a single member, CD1d. CD1 molecules are also found in many other mammals, including rats, guinea pigs, rabbits, and sheep (9) as well as in chickens(10). Evolutionarily, it appears that the CD1 cluster is the result of a gene duplication some 540 million years ago(11). Not knowing the full extent and diversity of the microbial lipid antigen collection, it is difficult to understand some unusual features of the CD1 gene evolution, such as the absence of the group 1 genes in the mouse, the multiplicity of CD1 genes in the guinea pig, or the emergence of CD1e in multiple species(12). It is likely that CD1 evolution and its selection under infectious pressure obeys the very same rules that govern classical MHC evolution with the caveat that CD1 genes are not in a hot spot of recombination/mutagenesis like MHC genes. However, the polymorphism found in mouse CD1d genes both in non-coding(13) and in coding regions of the gene(14) has functional consequences that support this general idea that selective pressure applies to CD1 genes as it does on MHC genes.
Structure of the CD1 Molecules
Similar to the paradigm shift created by the first structure determination of an MHC protein in 1987(15), the elucidation of the mouse CD1d structure in 1997 by Wilson’s group(2) was a turning point in our understanding of the functions of the protein. Two key observations came from this work: one expected, CD1 was structurally speaking an MHC class I and class II-like molecule, and one unexpected and paradigm changing: the revelation that the binding surface included two hydrophobic antigen-binding pockets, A’ and F’ (2). The A’ pocket was large, with a width of ~15Å and a length of ~20Å, whereas the F’ pocket was smaller with dimensions of 6–7Å by 18Å. The binding groove was very narrow, allowing interactions between side chains of residues of the opposite α helices that covered a large part of the A’ pocket and created a narrow entrance that connected to the F’ pocket through a long tunnel (2)(Figure 1). The occupancy and display of glycolipids by CD1d was revealed a few years later when the structures of both mouse and human CD1d molecules were determined in complex with α-galactosylceramide(16, 17). The A’ pocket was filled by the fatty acid, the F’ pocket with the sphingosine, and the top portal accommodated the neck of the ceramide and displayed the sugar moiety for T cell recognition. This general mode of binding and display by CD1d has been confirmed for various glycolipids and extends to type 2 CD1 members.
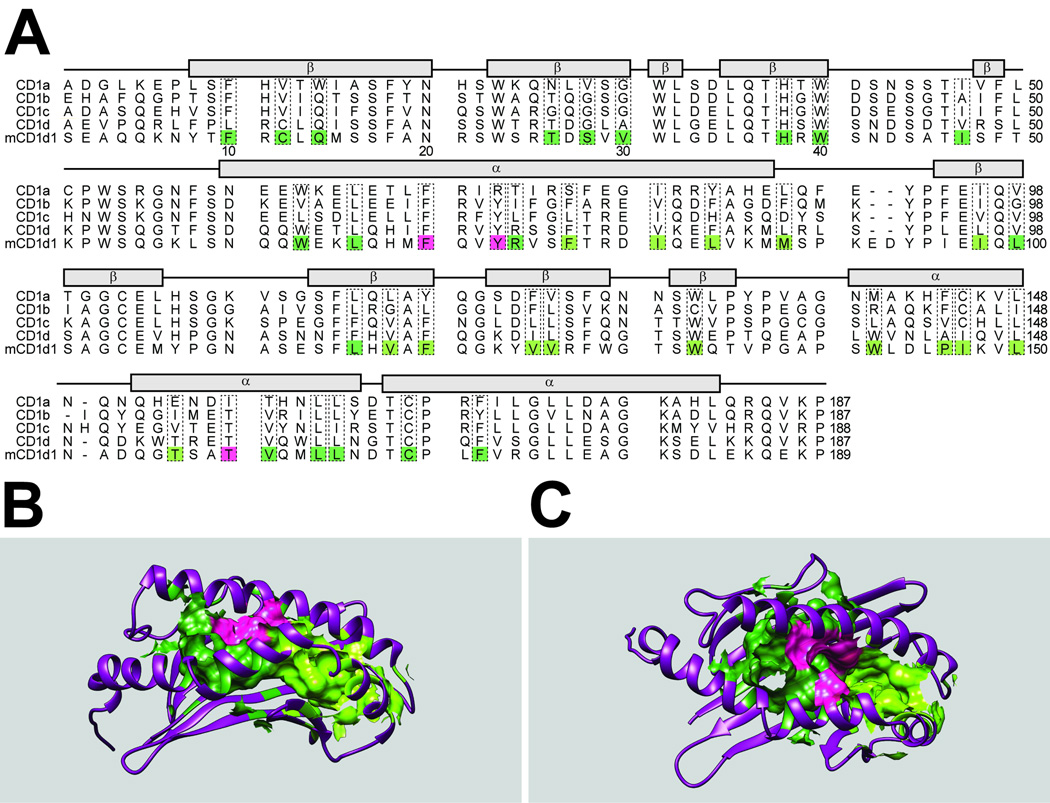
Figure 1. Comparison of the binding grooves of human and mouse CD1 isoforms.
A: Protein sequence comparison of the binding groove of human CD1a, b, c, d, and mouse CD1d1. All residues composing the surface of the binding groove are boxed with a dashed outline. In mCD1d1, residues composing the A’ pocket are boxed in dark green, residues composing the F’ pocket are boxed in light green, and residues composing the entrance portal are boxed in pink. Of note is the prevalence of hydrophobic residues that contribute to the binding groove surface.
B: Side view of the binding groove of mCD1d1 with the α1 chain in the foreground and the α2 chain in the background. Residues and their respective surfaces composing the A’ pocket are shaded dark green, residues and their respective surfaces composing the F’ pocket are shaded in light green, and residues and their respective surfaces composing the entrance portal are shaded pink (visualization was done with the Chimera software from UCSF(98) using pdb#1CD1.
C: Top view of the binding groove of mCD1d1 with the α1 on the bottom and α2 chain on top. Residues and surfaces are shaded as in Figure 1B.
In the case of CD1a, for which we determined the first crystal structure in complex with sulfatide(18), the arrangement of the binding groove and the display of the headgroup obeyed the very same rules as in the case of CD1d, with a sulfated galactose protruding from the center of the groove, easily accessible by anti-sulfatide T cells.
The size and complexity of the hydrophobic groove of CD1a, b, c, and d molecules vary to some extent and explain some diversity of antigen binding. CD1a has the smallest groove at ~1300Å3, followed by CD1d, ~1650Å3, CD1c, ~1800Å3, and CD1b, the largest and most complex of all with a 2200Å3 volume and two additional C’ and T’ pockets(19). Unique to CD1c is a D’ portal, an exit portal below the α1 helix, and an E’ portal, which connects the F’ pocket to the exterior (20). The sequence and structural differences between the binding pockets of human CD1a, b, c, d and murine CD1d1 are displayed in Figure 1.
The human CD1e crystal structure has also been determined and appears similar to the other members of the CD1 family. Its hydrophobic binding groove totals 2000 Å3 with a very large A’ pocket that can accommodate up to 30-carbon-long hydrophobic chains and an F’ pocket wider and more exposed compared to other CD1 molecules (21). However, no structure of CD1e bound to lipids has been shown yet, and the ability and role of CD1e to transfer lipids to other CD1 molecules remains very poorly understood at the molecular level.
Structure of the Ligands
A number of recurring features concerning the lipids bound to the various CD1 molecules have emerged from the small series of CD1-lipid complex structures that is now available. As mentioned above for α-galactosylceramide bound to CD1d, the overall disposition of glycerol-based lipids and glycosylceramides bound to the various CD1 molecules is very similar with the headgroup exposed and protruding at the center of the groove for direct TCR engagement, and the acyl chains occupying the A’ and F’ pockets. In the case of ceramide binding, the occupancy of the A’ and F’ pockets by the fatty acid chain and sphingosine chain, respectively, has always been observed. This particular orientation, and the interaction between the neck of the ceramide and CD1(16, 17) allows very distinct displays of the proximal sugar depending on its α- or β–linkage to the ceramide and the consequent differential recognition of anomers of monoglycosylceramides by B and T cells(22–24). In the case of diacylglycerol-based lipids, it has been shown that the A’ and F’ pockets could be alternatively filled by either of the two fatty acids orienting the head groups differently for T cell recognition and discrimination(25).
One often overlooked structural feature of CD1 molecules is the presence of a central pole in the A’ pocket that forces the alkyl chain to adopt a hooked conformation; such a conformation is naturally favored by the presence of unsaturation in the acyl chain of ceramides. One additional, recurring feature of many of the CD1-lipid structures that we have now observed is that the hydrophobic groove always attempts to be completely filled, and this was exemplified by the presence of a number of detergent molecules in the groove of CD1b(19). More relevant to biology, when alkyl chains are reduced in size, the presence of fatty acids of various lengths has been reported in the A’ pocket of both CD1d and CD1c-lipid complexes(16, 20) and found to help fill the groove. The origins and mode of loading of these fatty acids are still unknown. Their function for the association of CD1 molecules with lysolipids and presentation to T cells is most likely critical (see below for CD1a). These observations also lead to a consideration that could be relevant to clinical situations in which small hydrophobic drugs may bind to CD1 molecules and inhibit or modify binding and T cell recognition. This possibility is supported by the identification of stimulatory non-lipidic small molecules capable of binding to CD1d(26).
Some anomalies or new rules?
Very recently, a body of work centered on the identification of ligands for CD1a has yielded a series of “non-interfering” ligands that allowed T cell recognition of the roof of the A’ pocket without participation of the lipid itself(27). These permissive ligands included headless lipids such as squalene(28), as well as common lysophospholipids such as lysophosphatidylcholine, or common phospholipids such as phosphatidylcholine, phosphatidylglycerol, and phosphatidylinositol(28). The need to balance the presentation of CD1a itself, which could be defined as “real autoreactivity”, with the presentation of common lipids by CD1a is probably best illustrated by the response to bee venom and the production by the venom phospholipase A2 of a variety of ligands that include a mix of “non-interfering” and recognizable ligands with exposed headgroups(29).
Overall, it appears that CD1 molecules can bind all classes and varieties of lipid ligands(27, 30, 31). This diversity is not surprising given the characteristics of the CD1 groove. However, the physical properties of these lipids are extremely variable. While some reside in membranes and require extraction prior to binding to CD1, others, such as fatty acids, are reasonably soluble and capable of accessing the CD1 pockets directly. Therefore, the biology of ligand binding to CD1 must be necessarily complex and highly regulated to allow the immune surveillance of all lipids.
Synthesis and Trafficking of CD1 Molecules
The assembly and display of the CD1-lipid structures that we have just described offer some obvious challenges, as this process requires not only the presence of CD1 in a particular location but also a set of accessory molecules capable of extracting and/or transferring lipids, and the appropriate physicochemical environment to allow this process to occur. As we describe below, the key to CD1 function in antigen presentation is subcellular targeting.
Synthesis and Quality control
The beginning of the life of CD1 molecules is banal and very similar to MHC class I molecules. Our current knowledge has been gathered over many decades and mainly from studying murine CD1d molecules, but it appears that all group 1 and 2 CD1 molecules obey similar rules for synthesis and assembly in the endoplasmic reticulum (E.R.)(32) Very much like MHC class I molecules, CD1 molecules are folded and associated with β2-microglobulin in the E.R. following a succession of quality control checkpoints including CD1d heavy chain association with calnexin and calreticulin in a glycan-dependent fashion(33) and association with ERp57 to allow disulfide bond formation(34). After synthesis, CD1d heterodimers travel through the secretory pathway through the Golgi apparatus to reach the cell surface 1 to 2.5 hours post-translation (33, 34). Similarly to MHC class I molecules, it appears that the CD1 binding groove does not leave the E.R. empty but rather loaded with a lipid cargo. The nature of this cargo and its mode of loading are still debated, mainly because of the technical limitations in characterizing lipid-protein interactions in cells. Indeed, the repertoire of lipids bound to CD1 molecules cannot be studied using the techniques that have been developed for analyzing the peptide repertoire of MHC molecules. The use of detergents to access the cellular CD1 molecules will disrupt not only the CD1-lipid interaction but also solubilize a large number of cellular lipids that can then bind inside the CD1 groove in a competitive manner. Consequently, using these “classic” approaches, it is not possible to directly interrogate the nature of the lipidome of the E.R. CD1 molecules.
Indirect evidence has been reported that phospholipids were associated to CD1 molecules in the E.R. and that microsomal triglyceride transfer protein (MTP) may play a role in this loading(35). However, if it is clear that the biology of CD1 is profoundly affected upon deletion of MTP in both mouse(35, 36) and man(37), and that MTP is known to affect the lipid loading of Apolipoprotein B in the E.R., it is also obvious that MTP has a profound effect on lysosomal functions and the loading of CD1d molecules in late endosomal compartments(36). Similar issues have been discussed for all CD1 molecules(32).
In any instance, the way forward to study the important question of E.R. lipid loading resides in the development of new techniques in which CD1 molecules can be recovered without using detergents, or using detergents that are not capable of displacing the CD1-bound lipids, or through the production of antibodies or T cell receptor tetramers specific for particular CD1d-lipid complexes, which has been achieved for CD1d-α galactosylceramide(38).
Trafficking and targeting of CD1 molecules to lipid antigen processing compartments
The cellular lipid antigens that CD1 molecules survey are located in various organelles and compartments but all converge to the endosomal system for recycling and degradation. This location is also where most exogenous lipid antigens are directed for processing. Thus, CD1 biology is totally dependent on the capacity of CD1 molecules to survey the various subsets of endosomal vesicles to associate with relevant antigens. This is achieved differently by the four human CD1 molecules and the unique mouse CD1d member, using similar principles of subcellular trafficking: cytoplasmic targeting motifs. The demonstration of this principle was performed by deleting the cytoplasmic tail of CD1d, or the mutation of the critical tyrosine (RFKRQTSYQGVL) of this segment, both in cell lines and in vivo after gene knock in(39–41). This single tyrosine allows interaction with the adaptor protein AP-3(42, 43), directing CD1d to late endosomal, Lamp-1 positive compartments, where it accumulates(42, 43). It is important to mention that the deletion of the tail of CD1d did not impair cell surface expression but that it profoundly affected the presentation of both endogenous and exogenous lipid antigens(41), demonstrating the dependence of lipid presentation on the localization of the CD1 molecules to endosomal compartments.
CD1d also interacts with the invariant chain (Ii), the normal targeting chaperone of MHC class II molecules(39). Although this interaction has been shown for both mCD1d and human CD1a(44), the consequences of this association on the functions of CD1 molecules remain unclear. In the case of CD1d, Ii was shown to favor a direct targeting of CD1 from the trans-Golgi network to the lysosome but this pathway did not rescue CD1d presentation after deletion of the cytoplasmic tail(39, 41, 45). In invariant chain deficient mice, the phenotype was discrete, with an alteration of thymic selection of Vβ7 NKT cells and an inability of presenting antigens from Mycobacterium tuberculosis (46), indicating that some lipid antigens, including endogenous ligands, could only be loaded onto CD1 molecules in compartments where Ii could direct them. In the case of CD1a, Ii appeared to facilitate the cycling of CD1 from the surface to the early recycling endosome, but the functional benefits of this accessory function have not been evaluated, yet(44).
Human CD1 molecules
CD1a is the only human isoform to lack a tyrosine-based motif in its cytoplasmic tail(47). Consequently, it cannot traffic to the late endosomal system and is limited geographically to the early recycling compartments(44). As mentioned, Ii also plays a role in this distinctive trafficking of CD1a in the early endosome(44).
In contrast, CD1b traffics efficiently to the late endosomes and lysosomes(47, 48) by using a YXXZ cytoplasmic tail sequence, in which the tyrosine is spaced from a large hydrophobic residue by two amino acids. This motif allows the binding to both AP-2 and AP-3 adaptor proteins (48–50)
A similar motif allows CD1c to access the transferrin receptor positive early and medial endosomes, which express TfR (48), and later Lamp-1 positive compartments, giving this isotype the broadest distribution of all human CD1 molecules(51).
Human CD1d behaves very similarly to the mouse CD1d with an AP-3-mediated accumulation in late Lamp-1 positive endosomal/lysosomal compartments(49).
Finally, the fifth human CD1 isotype, CD1e, stands out due to its unique mode of trafficking. Before accessing the endosomal/lysosomal compartments during the maturation of dendritic cells and being cleaved, CD1e accumulates in the late Golgi and trans-Golgi network without ever accessing the cell surface. This behavior is controlled by the ubiquitination of the cytoplasmic tail of CD1e (52).
Overall, the synthesis, quality control, trafficking, and lipid loading of CD1 molecules described above, exemplified by CD1d, is summarized in Figure 2.
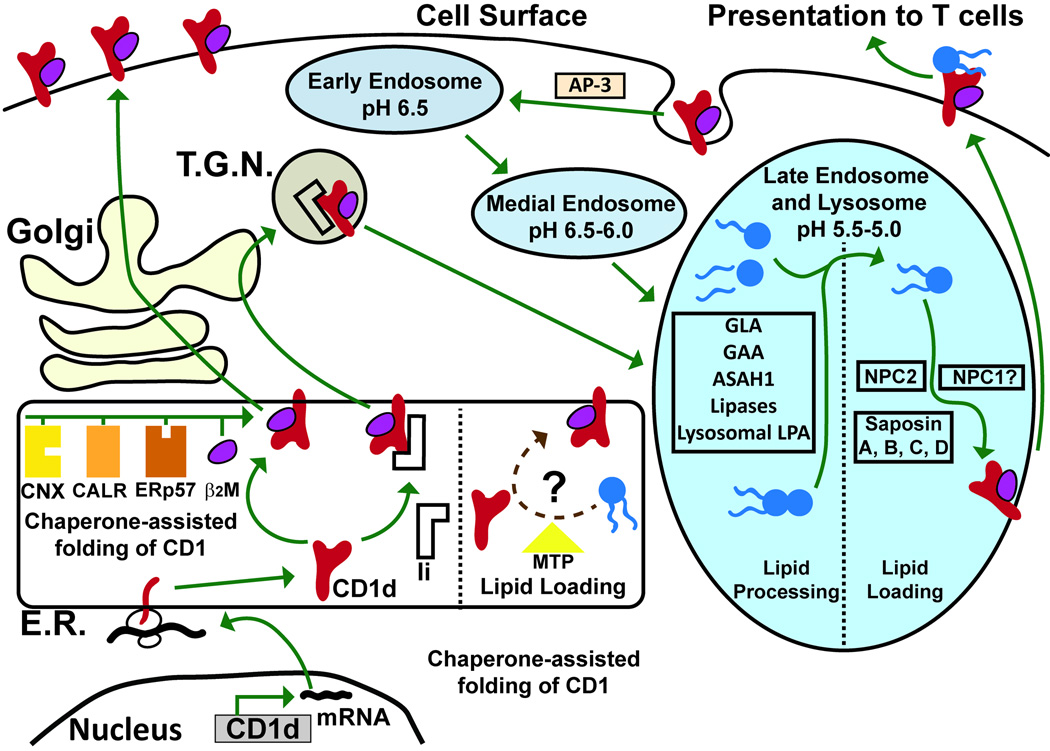
Figure 2. Cell biology of CD1d molecules.
First, the CD1d mRNA is translated into the endoplasmic reticulum (E.R.), where it undergoes chaperone-assisted folding via interaction with calnexin (CNX), calreticulin (CALR), and ERp57 until it forms a heterodimer with β2-microglobulin and associate or not with the invariant chain (Ii). Also in the E.R., microsomal triglyceride transfer protein (MTP) may play a role in loading some lipids to CD1d. After association with β2-microglobulin, CD1d is transported through the Golgi to the cell surface. It is internalized through interaction between its cytoplasmic tail and the AP-3 adaptor. This interaction and its disruption will allow trafficking through the endocytic pathway, and delivery to the late endosome and lysosome. Association with the invariant chain in the E.R. results in CD1d entering the trans-Golgi network, where it is directly transported to the late endosome and lysosome. In the late endosome and lysosome, lipid processing proteins alter lipids for pH-dependent loading to CD1d via NPC2, and saposins A, B, C, and D. The lipid-loaded CD1d then traffics to the cell surface, where it presents its lipid antigen to T cells.
Antigens Presented by CD1 Molecules
Without going through the exhaustive list of lipid antigens that can be presented by CD1 molecules and that have been highlighted in many reviews(27, 53), we will focus on six particular aspects of lipid biology that are relevant to CD1 antigen presentation.
Chemical diversity of lipid antigens
The examination of the diversity of peptides bound to MHC molecules using the now classic immunoprecipitation-elution-mass spectrometry sequence(54) has its own limitations linked to the loss of low affinity binders, the inherent affinity/avidity properties of the antibody used for pulldown, and the sensitivity of LC MS/MS detection techniques themselves. As a result it is estimated than no more than 30–50% of the “peptidome” associated to MHC is actually examined using the current technology. The situation is direr for studies aimed at analyzing the repertoire of lipids bound to CD1 molecules because they cannot be carried out in the presence of common detergents that would disrupt the CD1-lipid interaction. Nevertheless, the use of surface cleavable or/and soluble secreted CD1 molecules have allowed some limited studies. Those have delivered an important message: a majority of the most abundant phospholipids and glycolipids can associate with CD1 molecules(31, 32). This finding establishes a fundamental difference with MHC molecules and their peptide repertoire that is limited by the requirement for “binding motifs”.
Specificity of loading or no specificity?
This theoretical absence of discrimination of CD1 molecules for lipid binding does not mean that all lipids will be equally represented in the groove of CD1 molecules. Important chemical properties such as length and saturation of the acyl chains(16, 55, 56) will modify critical micelle concentrations, transport, and transfer into CD1 molecules, limiting the CD1-bound lipidome. The other important limiting factor to this lipid repertoire is the ability of each individual lipid to load into the groove of CD1 by itself (e.g. soluble short fatty acids), or to require assistance in this process for membrane extraction and then transfer (e.g. glycolipids). The localization of CD1 molecules to a subset of subcellular compartments should also be influential in repertoire selection since it is unlikely that all lipids and all accessory lipid loading molecules are present in all late endosomal compartments. This concept is best exemplified by the loading of gangliosides onto CD1 in compartments where gangliosides are degraded(57) and that are rich in particular lipids such as bis(monoacylglycero)phosphate (BMP, also called LBPA)(58, 59) which surprisingly are not known to be loaded onto CD1. In this case, it is most likely that the absence of loading is linked to the absence of a transfer protein capable of extracting LBPA from the membrane and transferring it to CD1, but this discretionary loading, despite the promiscuity of the CD1 binding pocket, remains unresolved.
Why the endosomal system for processing and loading?
As we have seen, CD1 molecules are localized to acidic compartments. In the mouse a single isoform patrols as much of the endosomal system as possible by using a targeting motif and the association with the invariant chain. In humans, the coverage is optimized by the existence of four isoforms with partial localization overlap and the association with the invariant chain. Similar to MHC class II molecules, CD1 localizes to the endosome/lysosome network because it is the converging compartment where exogenous antigens are brought for processing and endogenous antigens are recycled, giving the ability to CD1 molecules to sample both endogenous and exogenous lipids. In addition, because the late endosomal vesicles, especially multivesicular bodies(60), are where lipids are enzymatically degraded, CD1 can co-opt lipid-specific enzymes for its biology, namely antigen processing and antigen binding.
Exogenous versus endogenous antigens
Exogenous lipid antigens that can be loaded onto CD1 are not all microbial in origin. Indeed, a large number of self-lipids associated to lipoproteins are delivered constantly to antigen presenting cells and inside the endosomal system by receptor-mediated uptake. Similar to microbial lipids, these endocytosed lipids are destined to degradation and utilization as metabolic sources but can be “intercepted” and loaded onto CD1 molecules. Even though real endogenous lipids (defined as being synthesized by the cell itself and not having left that cell) can access the lysosomes through multiple pathways(61–63), it does not appear that their processing and loading are different from that of exogenous lipids. However, the details of endogenous CD1-binding lipid trafficking and the role of the major lipid transport proteins needed for non-vesicular transport have not yet been investigated(63, 64). Beyond the issues of trafficking between and within discrete subcellular compartments, the main issue that splits endogenous and exogenous antigens is our ability to determine their nature. In the case of exogenous microbial lipids, the easy access to large quantities of homogenous material makes structural determination much easier than in the case of cellular lipids. This situation explains why the nature of many relevant microbial glycolipids has been determined(5, 27, 65, 66) while so few endogenous antigens have been identified.
Anything unique about endogenous ligands?
As our knowledge stands today, within the small family of endogenous lipids that are presented by CD1 molecules, we can distinguish two branches. One of them is made of well-described lipids for which tissue distribution and functions are known such as squalenes in the skin that end up being presented by CD1a molecules(28), gangliosides such as GM2 that can be presented by CD1b(19), sulfatide, highly represented in the central nervous system and are presented by CD1a(18) as well as mouse and human CD1d molecules(67, 68), and phospholipids such as phosphatidylethanolamine(69) and phosphatidylglycerol(70), ubiquitous molecules that can be presented by either CD1c or CD1d. The frequency and potential homeostatic or pathological role that the recognition of these CD1-self-lipid complexes could lead to are still largely unknown. The second branch of the endogenous lipid family is shrouded in a different kind of mystery. While their functions are well known and consist of selecting in the thymus and activating in the periphery NKT cells, their origin, synthesis, distribution, modification, and alternate functions are largely unknown. They consist of α-linked monoglycosylceramides and were thought to be produced only by lower organisms such as bacteria(65, 71, 72), or porifera(73, 74), before being discovered in mammalian cells(22). One additional interesting characteristic of this family of endogenous ligands is that they have been functionally mimicked by non-ceramide based glycolipids such as α-glucosyl diacylglycerol as found in group B streptococci or Streptococcus pneumoniae, (75), or α-galactosyl diacylglycerol made by Borrelia burgdorferi (76).
The need for synthetic ligands and their limitations
In all cases, when an immunologically relevant lipid is isolated and characterized from a natural source, the confirmation of chemical nature, structure, and potency must be verified by chemical synthesis. However, technical limitations impede such studies. First, chemical synthesis of lipids and glycolipids is often challenging, especially when it comes to producing pure anomeric forms of the sugar head group. In addition, standard methods of detection such as mass spectrometry cannot distinguish anomers of sugars, and NMR requires relatively large amounts of the compound and thus lacks sensitivity. As a result of this difficult situation, investigators can be misguided and believe that one anomer is bioactive when, in fact, a contamination with the other anomer is responsible for the biological activity. Such a situation recently arose with the assignment of NKT cell stimulatory activity to β-glucosylceramide(77), before it was found that a contamination of the synthetic glycolipid was actually the active compound(22). To circumvent this important limitation of synthetic chemistry, we pioneered a simple assay that consists of using enzymes and/or lectins that are anomer-specific to eliminate or prove the existence of contaminants(22). This principle, which was used successfully to distinguish α and β-glucosylceramides using recombinant acid glucosylceramidase (GBA), an enzyme specific for β-glucosylceramide, has now been used by others(78), and expanded in our laboratory to the testing of the anomers of galactosylceramides using recombinant galactosylceramidase (GALC)(L.T., unpublished).
Antigen Processing and loading onto CD1 molecules
The succession of events that will lead to the display of the lipids we have just discussed by cell surface CD1 molecules is very similar to the processing and presentation of peptides by MHC class II molecules. Indeed, both CD1 and MHC class II molecules have to come to the same late endosomal compartments to pick up ligands before they are fully degraded and used for basic metabolic purpose. The competition between proteolysis and MHC loading is still incompletely understood, and many of the same questions apply to CD1 presentation. The three main classes of CD1 antigen presenting cells are B cells, dendritic cells, and thymic T cells; the endocytic capacities of all three cell types are very different. Double positive thymocytes expressing CD1 primarily present endogenous lipids for the selection of CD1-restricted T cells and do not have to be efficient at endocytosis(4). B cells and dendritic cells use phagocytosis and receptor-mediated endocytosis to capture whole or fragments of microorganisms, respectively, in order to deliver lipids to CD1-containing subcellular compartments, and these uptake capacities are heavily influenced by the differentiation stage of the cell(79). An additional mode of delivery of lipids to antigen presenting cells relies on lipoprotein transport. Indeed, a large number of lipids traffic through organisms by using the packaging of lipoproteins, and this aspect is not only relevant to microbial lipids but also to all immunologically active lipids that would be administered to patients. This mode of uptake/delivery was exemplified for α-galactosylceramide that was shown to associate to apolipoprotein E within high density and very low density lipoproteins (LDL) to be taken up by cells through LDL receptor-mediated endocytosis(80) as well as scavenger receptor capture(81). In addition, it was also demonstrated that protein-mediated non-lipoprotein lipid transfer was important for the biological activity of α-galactosylceramide and derivatives(82).
Antigen processing
The principle of antigen processing for CD1 presentation was established first by using synthetic derivatives of α-galactosylceramide(83). For instance, it was shown that Gal(α1–2)-α-galactosylceramide could become stimulatory only after the hydrolysis of the additional galactoses by α-galactosidase(83). This observation has now been expanded to natural glycolipids such as the α-galactopyranosyl(1→2)-α-glucosyl diacylglycerol of Streptococcus pneumoniae (75) that also requires the removal of the distal galactose to become stimulatory, or phosphatidylinositol mannosides of Mycobacterium tuberculosis that require trimming of the mannose tree by acidic α mannosidase to be presented by CD1b and seen by T cells(84). The hydrolysis of the headgroup of a glycolipid can also lead to its inactivation. This processing has been shown to be critical for the inactivation of the endogenous α-galactosylceramide and is mediated by acid α galactosidase (GLA)(22, 85).
The acyl chains of lipids are also modified to allow loading and/or stimulatory activity of particular lipids. Lysosomal phospholipase A2 (LPLA2), an endocytotic compartment-localized enzyme which cleaves the acyl chain of various phospholipids appears to be important for the presentation of certain endogenous ligands in vivo and the processing of exogenous ligands such as galactosyl-α1–2-galactosyl ceramide in vitro and their presentation to NKT cells(86). Similarly, we have shown that acid ceramidase was important to degrade endogenous α-galactosylceramide and produce α-lysogalactosylceramide, a substrate of GLA(22).
Mechanisms of lipid loading onto CD1 molecules
It was conceptually obvious that lipid loading onto CD1 molecules needed the assistance of lipid transfer proteins (LTP) since CD1a, b, c, and d are not capable of extracting lipids from membranes themselves. While MHC class II has co-evolved a MHC-class II molecule that assists in peptide loading, CD1 members have simply co-opted the molecules that are essential to lipid and glycolipid processing and metabolism in the lysosomes. The number of lysosomal lipid transfer proteins is very small and has allowed an easy evaluation of each of them in CD1 biology. Saposins A, B, C, and D, derived from prosaposin by proteolytic cleavage upon secretion/uptake by antigen presenting cells, appear to be the most important lipid transfer proteins for the loading and exchange of lipid within the groove of CD1 molecules(87–89). Saposins are small (8–11 kDa (90)) proteins necessary for glycolipid (91) and sphingolipid (90) degradation in the endosome. They function by non-enzymatic mechanisms and only at acidic pHs. Structural work as well as lipid exchange experiments using liposomes have shown that they use a tweezer-like mechanism to extract lipids from membranes by inserting, around the headgroup of phospho- and glycolipids, hydrophobic residues exposed at the tip of surface loops(92). Each saposin has preferences for a subset of lipids, based on the nature and charge of the headgroup but specificities are largely overlapping. Similar to saposins, Gm2A, another lysosomal lipid transfer protein needed for the enzymatic degradation of ganglioside GM2, was shown also to being capable of loading antigens to CD1d (87). In addition, saposins and Gm2A were shown to unload lipids bound to CD1d (87), supporting a “tug-of-war” model for antigen loading and exchange in which the LTPs will either load or extract lipid from CD1 based on the affinity of both proteins for the same lipid(87). The Niemann-Pick type C1 and C2 proteins (NPC1 and NPC2, respectively) have also been shown to be important for antigen loading to CD1d. While NPC2 was shown to function like saposins for the editing of endogenous lysosomal lipid ligands(93), NPC1 appeared to play an indirect role by interfering with the proper late endosome-lysosome fusion(94). There is currently no structural data supporting the hypothesis of the “tug of war” of lipid editing onto CD1 molecules.
In humans, an additional LTP has been shown to be important for the loading of microbial lipids to CD1b: the cleaved soluble form of CD1e(84).
Once loading is completed, the CD1 molecule can be transported to the cell surface. The nature of the interactions that retain non-loaded or partially loaded molecules inside the lysosome, or that inversely allow transport to the surface, are unknown.
Concluding remarks: analytical methods for lipids, limitations
If the overall operational mode of lipid processing and loading onto CD1 molecules in the lysosome is reasonably well understood, there is no question that further studies are limited by the available techniques. Over the past decade, mass spectrometry (MS) has taken over the classical biochemistry approach to studying lipids. However, MS does not offer the sensitivity and analytical power that one might need to discover minor and/or unknown species. The utilization of techniques such as multiple reaction monitoring MS (MRM MS) can address sensitivity issues but they require knowledge of the spectra of the sought compounds and the libraries of profiles for lipids and glycolipids are still very poorly populated (95). Currently, the most complete database, “LIPID MAPS”, features about 40,000 lipids. In addition, MS is inherently plagued with its inability to distinguish anomeric sugars and subtle regiochemistry.
As it stands, the combination of classic thin-layer chromatography or liquid chromatography with MS-MS methods and MRM is the most sensitive approach to study non-radioactively labeled lipids(96). Radiolabeling offers the great advantage of sensitivity but it is costly and cumbersome, and has been used less and less. At the other end of the spectrum, NMR spectroscopy gives the most definitive and accurate structural information, but it remains limited by the relatively large amounts of material it requires(97). Finally, it is noteworthy to mention that immunologists have underused one of their most powerful tools for structural determination and discrimination of lipids: antibodies. Anti-lipid antibodies have been used extensively in the field of neuroscience but rarely in immunology. There are still very few anti-lipid and glycolipids antibodies available but their sensitivity and ability to distinguish discrete chemical variations is unsurpassed(22, 58) and could yield interesting insights into the biology of antigen presentation by CD1 molecules.
Acknowledgments
This work was supported by NIH grant (1RO1 AI102892 to L.T.).
Footnotes
Conflict of interest statement:
I declare that none of the authors of the manuscript entitled ‘The processing and presentation of lipids and glycolipids to the immune system’ have a conflict of interest with respect its the content.
Bibliography
- 1.Castano AR, et al. Peptide binding and presentation by mouse CD1. Science. 1995;269:223–226. doi: 10.1126/science.7542403. [DOI] [PubMed] [Google Scholar]
- 2.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 3.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691–694. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 4.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annual review of immunology. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- 5.Van Rhijn I, Ly D, Moody DB. CD1a, CD1b, and CD1c in immunity against mycobacteria. Adv Exp Med Biol. 2013;783:181–197. doi: 10.1007/978-1-4614-6111-1_10. [DOI] [PubMed] [Google Scholar]
- 6.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and co-exists with TL in the same thymus. EMBO J. 1988;7:3081–3086. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–292. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 9.Porcelli SA, Modlin RL. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 10.Salomonsen J, et al. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc Natl Acad Sci U S A. 2005;102:8668–8673. doi: 10.1073/pnas.0409213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kasahara M. New insights into the genomic organization and origin of the major histocompatibility complex: role of chromosomal (genome) duplication in the emergence of the adaptive immune system. Hereditas. 1997;127:59–65. doi: 10.1111/j.1601-5223.1997.t01-1-00059.x. [DOI] [PubMed] [Google Scholar]
- 12.Dascher CC, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–5488. [PubMed] [Google Scholar]
- 13.Borg ZD, et al. Polymorphisms in the CD1d promoter that regulate CD1d gene expression are associated with impaired NKT cell development. J Immunol. 2014;192:189–199. doi: 10.4049/jimmunol.1301451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zimmer MI, et al. Polymorphisms in CD1d affect antigen presentation and the activation of CD1d-restricted T cells. Proc Natl Acad Sci U S A. 2009;106:1909–1914. doi: 10.1073/pnas.0808476106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. Structure of the human class I histocompatibility antigen, HLA-A2. Nature. 1987;329:506–512. doi: 10.1038/329506a0. [DOI] [PubMed] [Google Scholar]
- 16.Zajonc DM, et al. Structure and function of a potent agonist for the semi-invariant natural killer T cell receptor. Nat Immunol. 2005;6:810–818. doi: 10.1038/ni1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koch M, et al. The crystal structure of human CD1d with and without alpha-galactosylceramide. Nat Immunol. 2005;6:819–826. doi: 10.1038/ni1225. [DOI] [PubMed] [Google Scholar]
- 18.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 19.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 20.Scharf L, et al. The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Alles LF, et al. Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc Natl Acad Sci U S A. 2011;108:13230–13235. doi: 10.1073/pnas.1105627108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kain L, et al. The identification of the endogenous ligands of natural killer T cells reveals the presence of mammalian alpha-linked glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu ED, et al. Structural Basis for the Recognition of C20:2-alphaGalCer by the Invariant Natural Killer T Cell Receptor-like Antibody L363. The Journal of biological chemistry. 2012;287:1269–1278. doi: 10.1074/jbc.M111.308783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott-Browne JP, et al. Germline-encoded recognition of diverse glycolipids by natural killer T cells. Nat Immunol. 2007;8:1105–1113. doi: 10.1038/ni1510. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, et al. Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. P Natl Acad Sci USA. 2010;107:1535–1540. doi: 10.1073/pnas.0909479107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Rhijn I, et al. CD1d-restricted T cell activation by nonlipidic small molecules. Proc Natl Acad Sci U S A. 2004;101:13578–13583. doi: 10.1073/pnas.0402838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small-molecule display by CD1 and MR1. Nat Rev Immunol. 2015;15:643–654. doi: 10.1038/nri3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Jong A, et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol. 2014;15:177–185. doi: 10.1038/ni.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bourgeois EA, et al. Bee venom processes human skin lipids for presentation by CD1a. J Exp Med. 2015;212:149–163. doi: 10.1084/jem.20141505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 31.Cox D, et al. Determination of cellular lipids bound to human CD1d molecules. PLoS One. 2009;4:e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park JJ, et al. Lipid-protein interactions: biosynthetic assembly of CD1 with lipids in the endoplasmic reticulum is evolutionarily conserved. Proc Natl Acad Sci U S A. 2004;101:1022–1026. doi: 10.1073/pnas.0307847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim HS, Garcia J, Exley M, Johnson KW, Balk SP, Blumberg RS. Biochemical characterization of CD1d expression in the absence of beta2-microglobulin. J Biol Chem. 1999;274:9289–9295. doi: 10.1074/jbc.274.14.9289. [DOI] [PubMed] [Google Scholar]
- 34.Kang SJ, Cresswell P. Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J Biol Chem. 2002;277:44838–44844. doi: 10.1074/jbc.M207831200. [DOI] [PubMed] [Google Scholar]
- 35.Dougan SK, et al. Microsomal triglyceride transfer protein lipidation and control of CD1d on antigen-presenting cells. J Exp Med. 2005;202:529–539. doi: 10.1084/jem.20050183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sagiv Y, et al. A distal effect of microsomal triglyceride transfer protein deficiency on the lysosomal recycling of CD1d. J Exp Med. 2007;204:921–928. doi: 10.1084/jem.20061568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zeissig S, et al. Primary deficiency of microsomal triglyceride transfer protein in human abetalipoproteinemia is associated with loss of CD1 function. J Clin Invest. 2010;120:2889–2899. doi: 10.1172/JCI42703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salio M, et al. Modulation of human natural killer T cell ligands on TLR-mediated antigen-presenting cell activation. Proc Natl Acad Sci U S A. 2007;104:20490–20495. doi: 10.1073/pnas.0710145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 40.Rodionov DG, Nordeng TW, Pedersen K, Balk SP, Bakke O. A critical tyrosine residue in the cytoplasmic tail is important for CD1d internalization but not for its basolateral sorting in MDCK cells. J Immunol. 1999;162:1488–1495. [PubMed] [Google Scholar]
- 41.Chiu YH, et al. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 42.Elewaut D, et al. The adaptor protein AP-3 is required for CD1d-mediated antigen presentation of glycosphingolipids and development of Valpha14i NKT cells. J Exp Med. 2003;198:1133–1146. doi: 10.1084/jem.20030143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cernadas M, et al. Lysosomal localization of murine CD1d mediated by AP-3 is necessary for NK T cell development. J Immunol. 2003;171:4149–4155. doi: 10.4049/jimmunol.171.8.4149. [DOI] [PubMed] [Google Scholar]
- 44.Sloma I, et al. Regulation of CD1a surface expression and antigen presentation by invariant chain and lipid rafts. J Immunol. 2008;180:980–987. doi: 10.4049/jimmunol.180.2.980. [DOI] [PubMed] [Google Scholar]
- 45.Chiu YH, et al. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–110. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sille FC, et al. Critical role for invariant chain in CD1d-mediated selection and maturation of Valpha14-invariant NKT cells. Immunol Lett. 2011;139:33–41. doi: 10.1016/j.imlet.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugita M, et al. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–752. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 48.Briken V, Jackman RM, Watts GF, Rogers RA, Porcelli SA. Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. J Exp Med. 2000;192:281–288. doi: 10.1084/jem.192.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity. 2002;16:697–706. doi: 10.1016/s1074-7613(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 50.Briken V, Jackman RM, Dasgupta S, Hoening S, Porcelli SA. Intracellular trafficking pathway of newly synthesized CD1b molecules. EMBO J. 2002;21:825–834. doi: 10.1093/emboj/21.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugita M, van Der Wel N, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci U S A. 2000;97:8445–8450. doi: 10.1073/pnas.150236797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Angenieux C, et al. The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic. 2005;6:286–302. doi: 10.1111/j.1600-0854.2005.00272.x. [DOI] [PubMed] [Google Scholar]
- 53.Zajonc DM, Wilson IA. Architecture of CD1 proteins. Curr Top Microbiol Immunol. 2007;314:27–50. doi: 10.1007/978-3-540-69511-0_2. [DOI] [PubMed] [Google Scholar]
- 54.Stern LJ. Characterizing MHC-associated peptides by mass spectrometry. J Immunol. 2007;179:2667–2668. doi: 10.4049/jimmunol.179.5.2667. [DOI] [PubMed] [Google Scholar]
- 55.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 56.McCarthy C, et al. The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med. 2007;204:1131–1144. doi: 10.1084/jem.20062342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandhoff K, Kolter T. Topology of glycosphingolipid degradation. Trends Cell Biol. 1996;6:98–103. doi: 10.1016/0962-8924(96)80999-8. [DOI] [PubMed] [Google Scholar]
- 58.Matsuo H, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 59.Gruenberg J. Lipids in endocytic membrane transport and sorting. Curr Opin Cell Biol. 2003;15:382–388. doi: 10.1016/s0955-0674(03)00078-4. [DOI] [PubMed] [Google Scholar]
- 60.Schulze H, Kolter T, Sandhoff K. Principles of lysosomal membrane degradation: Cellular topology and biochemistry of lysosomal lipid degradation. Biochim Biophys Acta. 2009;1793:674–683. doi: 10.1016/j.bbamcr.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 61.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 62.D'Angelo G, et al. Vesicular and non-vesicular transport feed distinct glycosylation pathways in the Golgi. Nature. 2013;501:116–120. doi: 10.1038/nature12423. [DOI] [PubMed] [Google Scholar]
- 63.Lauria I, et al. GLTP mediated non-vesicular GM1 transport between native membranes. PLoS One. 2013;8:e59871. doi: 10.1371/journal.pone.0059871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanada K, et al. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 65.Mattner J, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 66.Gumperz JE, Brenner MB. CD1-specific T cells in microbial immunity. Current opinion in immunology. 2001;13:471–478. doi: 10.1016/s0952-7915(00)00243-0. [DOI] [PubMed] [Google Scholar]
- 67.Luoma AM, et al. Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity. 2013;39:1032–1042. doi: 10.1016/j.immuni.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zajonc DM, et al. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gumperz JE, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–221. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 70.Van Rhijn I, et al. Human autoreactive T cells recognize CD1b and phospholipids. Proc Natl Acad Sci U S A. 2016;113:380–385. doi: 10.1073/pnas.1520947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinjo Y, et al. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature. 2005;434:520–525. doi: 10.1038/nature03407. [DOI] [PubMed] [Google Scholar]
- 72.Wieland Brown LC, et al. Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS Biol. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 74.Costantino V, Fattorusso E, Imperatore C, Mangoni A, Freigang S, Teyton L. Corrugoside, a new immunostimulatory alpha-galactoglycosphingolipid from the marine sponge Axinella corrugata. Bioorg Med Chem. 2008;16:2077–2085. doi: 10.1016/j.bmc.2007.10.098. [DOI] [PubMed] [Google Scholar]
- 75.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kinjo Y, et al. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978–986. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- 77.Brennan PJ, et al. Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol. 2011;12:1202–1211. doi: 10.1038/ni.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brennan PJ, et al. Activation of iNKT cells by a distinct constituent of the endogenous glucosylceramide fraction. Proc Natl Acad Sci U S A. 2014;111:13433–13438. doi: 10.1073/pnas.1415357111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cao X, et al. CD1 molecules efficiently present antigen in immature dendritic cells and traffic independently of MHC class II during dendritic cell maturation. J Immunol. 2002;169:4770–4777. doi: 10.4049/jimmunol.169.9.4770. [DOI] [PubMed] [Google Scholar]
- 80.van den Elzen P, et al. Apolipoprotein-mediated pathways of lipid antigen presentation. Nature. 2005;437:906–910. doi: 10.1038/nature04001. [DOI] [PubMed] [Google Scholar]
- 81.Freigang S, et al. Scavenger receptors target glycolipids for natural killer T cell activation. J Clin Invest. 2012;122:3943–3954. doi: 10.1172/JCI62267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freigang S, et al. Fatty acid amide hydrolase shapes NKT cell responses by influencing the serum transport of lipid antigen in mice. The Journal of clinical investigation. 2010;120:1873–1884. doi: 10.1172/JCI40451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Prigozy TI, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291:664–667. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 84.de la Salle H, et al. Assistance of microbial glycolipid antigen processing by CD1e. Science. 2005;310:1321–1324. doi: 10.1126/science.1115301. [DOI] [PubMed] [Google Scholar]
- 85.Darmoise A, et al. Lysosomal alpha-galactosidase controls the generation of self lipid antigens for natural killer T cells. Immunity. 2010;33:216–228. doi: 10.1016/j.immuni.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paduraru C, et al. Role for lysosomal phospholipase A2 in iNKT cell-mediated CD1d recognition. Proc Natl Acad Sci U S A. 2013;110:5097–5102. doi: 10.1073/pnas.1302923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou D, et al. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Winau F, et al. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 89.Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 90.Kolter T, Sandhoff K. Principles of lysosomal membrane digestion: stimulation of sphingolipid degradation by sphingolipid activator proteins and anionic lysosomal lipids. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 91.Sandhoff K, Kolter T, Van Echten-Deckert G. Sphingolipid metabolism - Sphingoid analogs, sphingolipid activator proteins, and the pathology of the cell. Ann Ny Acad Sci. 1998;845:139–151. doi: 10.1111/j.1749-6632.1998.tb09667.x. [DOI] [PubMed] [Google Scholar]
- 92.Alattia JR, Shaw JE, Yip CM, Prive GG. Direct Visualization of Saposin Remodelling of Lipid Bilayers. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 93.Schrantz N, Sagiv Y, Liu Y, Savage PB, Bendelac A, Teyton L. The Niemann-Pick type C2 protein loads isoglobotrihexosylceramide onto CD1d molecules and contributes to the thymic selection of NKT cells. The Journal of experimental medicine. 2007;204:841–852. doi: 10.1084/jem.20061562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sagiv Y, et al. Cutting edge: impaired glycosphingolipid trafficking and NKT cell development in mice lacking Niemann-Pick type C1 protein. J Immunol. 2006;177:26–30. doi: 10.4049/jimmunol.177.1.26. [DOI] [PubMed] [Google Scholar]
- 95.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A. 2015;112:12549–12550. doi: 10.1073/pnas.1516878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 97.Gasa S, Nakamura M, Makita A, Ikura M, Hikichi K. Complete structural analysis of globoseries glycolipids by two-dimensional nuclear magnetic resonance. Eur J Biochem. 1986;155:603–611. doi: 10.1111/j.1432-1033.1986.tb09531.x. [DOI] [PubMed] [Google Scholar]
- 98.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]