Figure 2.
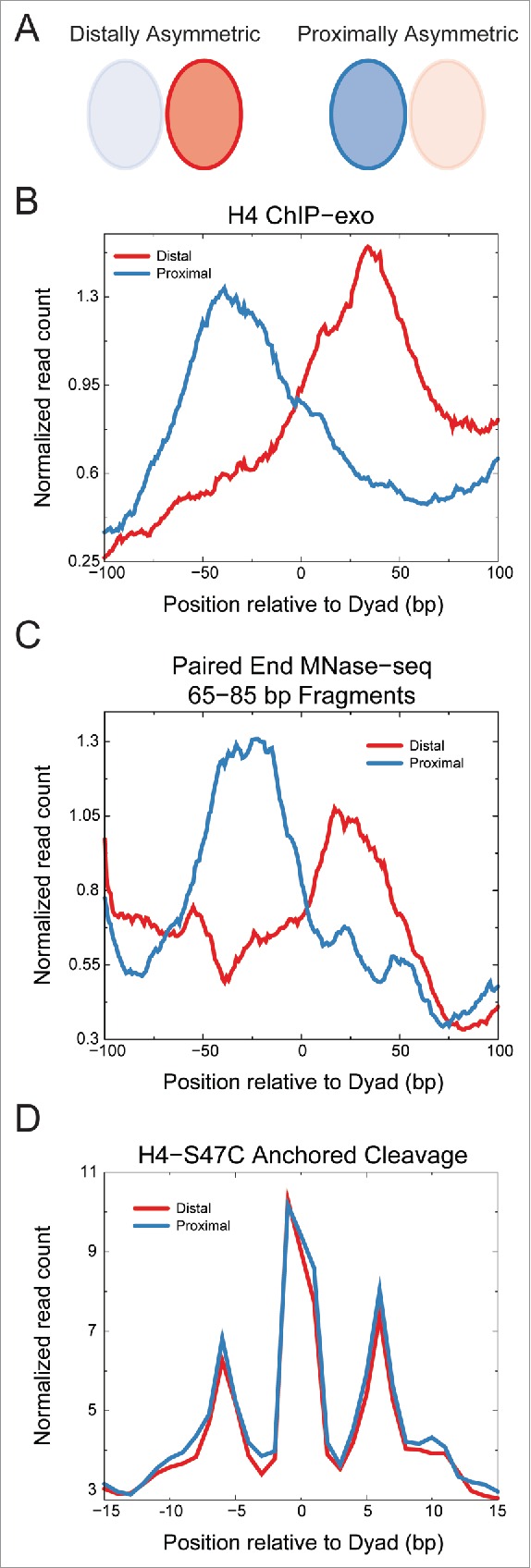
Asymmetric nucleosome positions identified by ChIP-exo show symmetric H4S47C-anchored cleavages. A) Schematic of ChIP-exo asymmetry relative to the transcription start site (TSS). Ratio of H4-ChIP-exo counts between proximal and distal sides of the dyad axis relative to the TSS from Rhee et al.20 was used to identify asymmetric nucleosome positions. The H4-ChIP-exo ratios were obtained from Table S4 of Rhee et al. and a cutoff of greater than 2 or less than 0.5 was used. B) Averages of H4 ChIP-exo data for the distally (n = 264) and proximally (n = 293) asymmetric nucleosome positions. The running average over a 15 bp window is plotted. C) Averages of centers of fragments from MNase-seq30 are plotted for distally and proximally asymmetric nucleosome positions identified by H4-ChIP-exo. The running average over a 15 bp window is plotted for fragments of 65–85 bp in length, which would include half-nucleosome protections. Enrichment is observed on the distal side of the dyad for the distally asymmetric positions and on the proximal side of the dyad for the proximally asymmetric positions. D) Average read counts of the ends of fragments generated by H4S47C-anchored cleavage16,18 are plotted for distally and proximally asymmetric nucleosome positions identified by H4-ChIP-exo. We observe no asymmetry in the H4S47C-anchored cleavage data for both distally and proximally asymmetric positions identified by H4-ChIP-exo.
