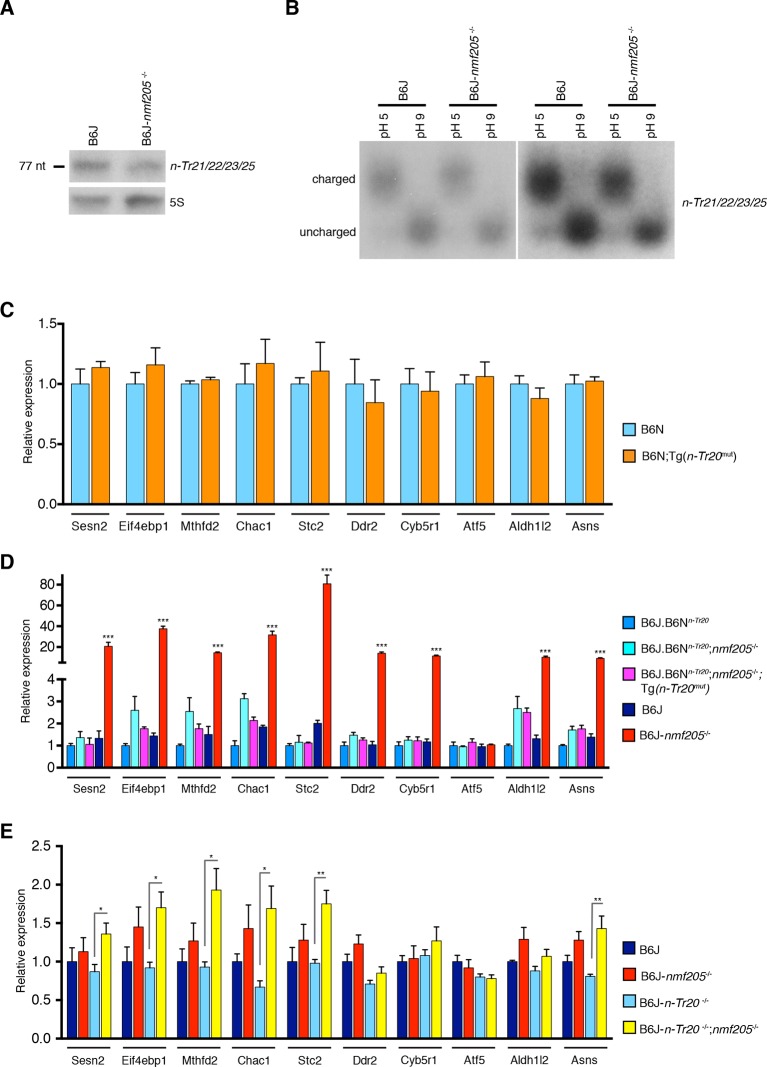
Figure 4. GCN2 activation in the B6J-Gtpbp2nmf205-/- brain is independent of uncharged tRNAArgUCU and unprocessed n-Tr20 tRNA.
(A) Northern blot analysis of cerebellar RNA from 3-week-old B6J and B6J-Gtpbp2nmf205-/- (B6J-nmf205-/-) mice using pooled probes to n-Tr21/22/23/25 tRNAs to assess the expression levels of the tRNAArgUCU isodecoder family. 5S was used as internal control. (B) Charged (pH 5) and uncharged (pH 9) tRNAArgUCU levels in the 3-week-old B6J and B6J-Gtpbp2nmf205-/- cerebellum. Note that the levels of uncharged tRNAArgUCU are negligible in both the B6J-Gtpbp2nmf205-/- and B6J cerebellum. A short (right) and a longer (left) exposure are shown. (C–E) RT-qPCR analysis of expression of ATF4 target genes. (C) Overexpression of the mutant n-Tr20 tRNA does not change expression of ATF4 targets in the 3-week-old cerebellum although increased levels of unprocessed n-Tr20 are present. (n=3 mice per genotype) (D) Overexpression of the mutant n-Tr20 tRNA does not change expression of ATF4 targets even in the Gtpbp2nmf205-/- mutant cerebellum at 3 weeks of age. (n=3) (E) ATF4 targets are significantly upregulated in the P0 brain of B6J-n-Tr20-/-; Gtpbp2nmf205-/- mice, although no unprocessed or uncharged forms of n-Tr20 are present. (n=4 mice per genotype) Error bars = SEM. *p<0.05, **p<0.01, and ***p<0.001 (Student's unpaired two-tailed t tests, C, E; one-way ANOVA, D).
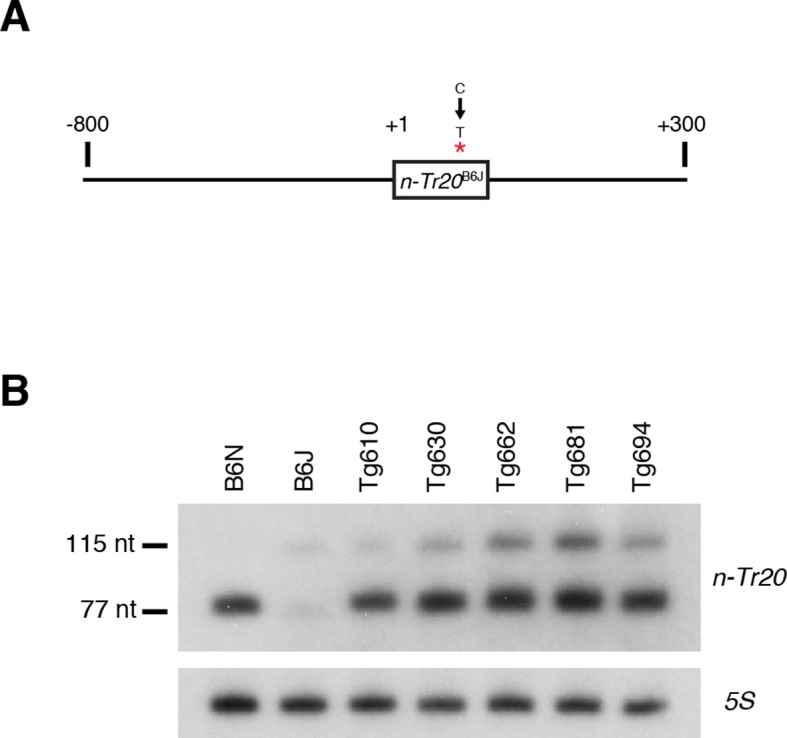
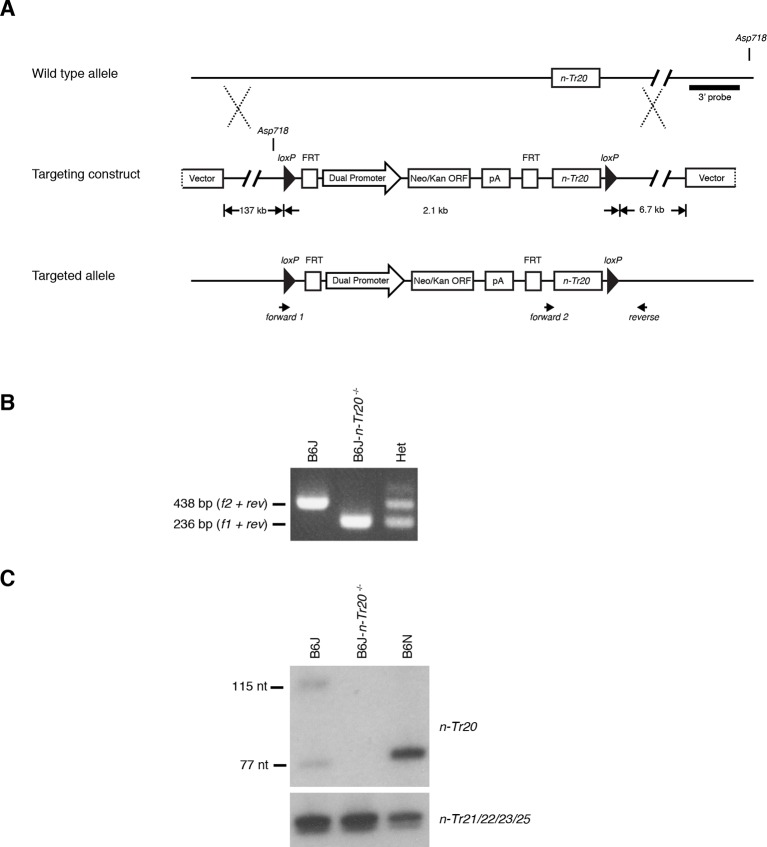
Figure 4—figure supplement 1. Generation of mice overexpressing mutant n-Tr20.