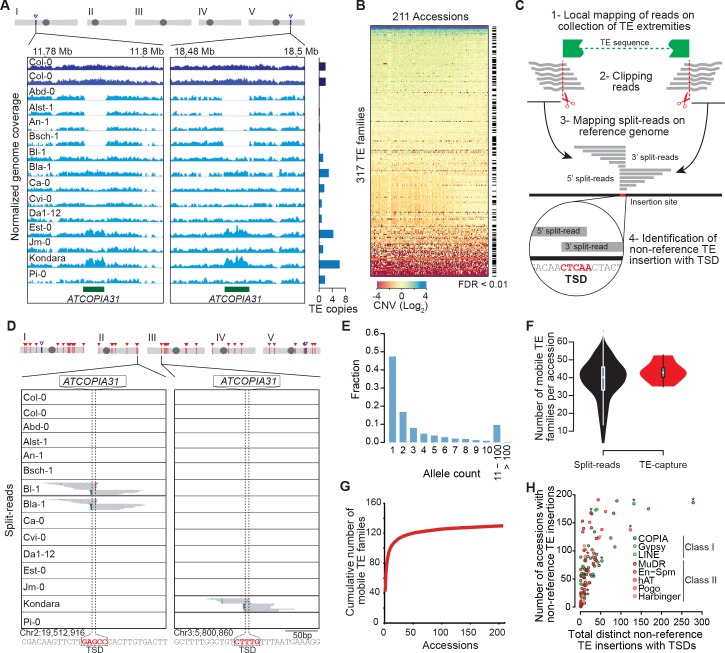
Figure 1. Overview of the A. thaliana mobilome.
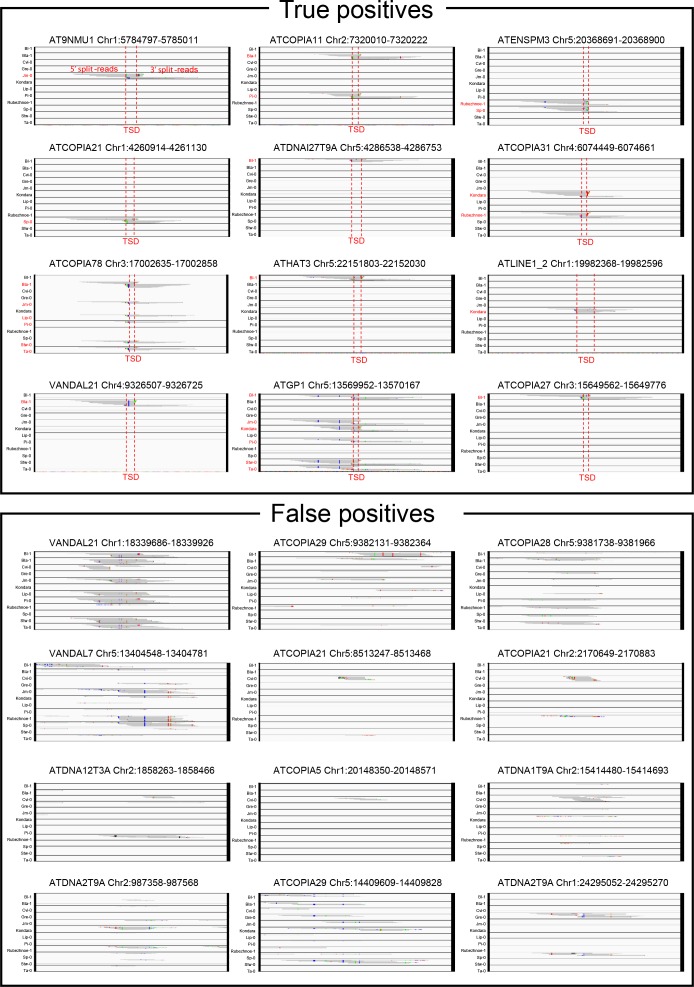
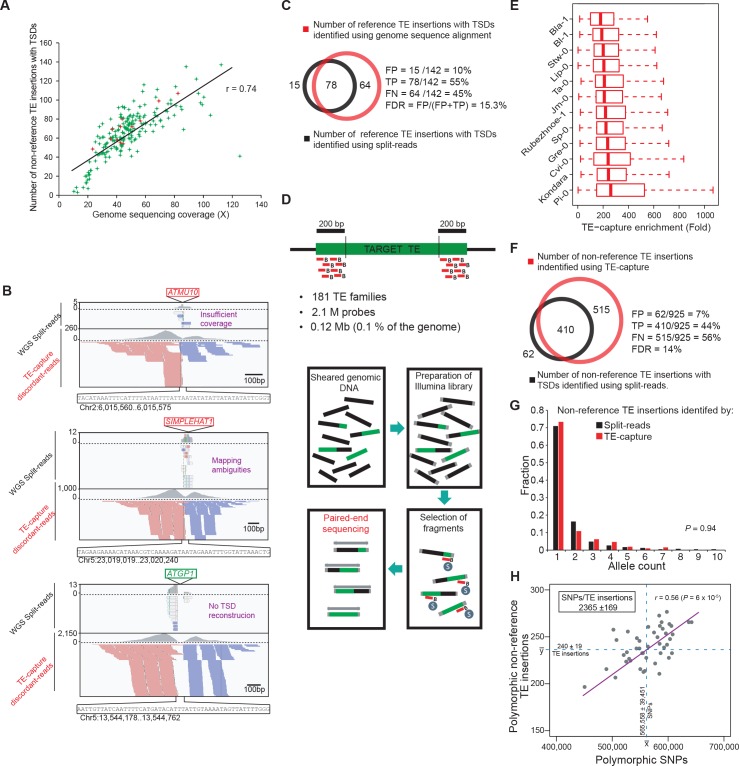
(A) Genome browser tracks showing normalized sequencing coverage over the two full-length ATCOPIA31 elements annotated in the reference genome (Col-0). CNV is detected as increased or decreased coverage in other accessions. Number of copies is indicated on the right. (B) Heat map representing CNVs (log2 ratio) for 317 TE families and 211 A. thaliana accessions. TE families with statistically significant CNV in at least one accession are indicated. Figure 1—source data 1 contains absolute copy number estimation of TE sequences. (C) Schematic representation of the bioinformatics pipeline to identify non-reference TE insertions with TSD using split-reads. 1- Reads are mapped on a collection of TE extremities from annotated TE sequences and reference sequences (Repbase update). 2- Reads aligning partially over TE extremities are extracted and clipped. 3- The unmapped portion of these split-reads are re-mapped on the Arabidopsis reference genome. 4- Non-reference TE insertions with TSDs are identified by searching for overlapping clusters of 5’ and 3’ split-reads. (D) Genome browser tracks showing split-reads for two non-reference ATCOPIA31 insertions and TSD reconstruction. Figure 1—source data 2 contains the coordinates of all non-reference TE insertions with TSDs. (E) Distribution frequency of allele counts for non-reference TE insertions with TSDs. (F) Number of mobile TE families per accession identified using split-read and TE-sequence capture. (G) Cumulative plot of the number of mobile TE families detected with increasing numbers of accessions. (H) The total number of non-reference TE insertions with TSDs is indicated in relation to the number of accessions with such insertions, for each of the 131 mobile TE families. Asterisks indicate the nine TE families with experimental evidence of transposition (Ito and Kakutani, 2014; Tsay et al., 1993). Figure 1—source data 3 contains the total number of distinct non-reference TE insertions with TSD for each TE family and super-family. Figure 1—figure supplement 2 shows TE-capture results. Figure 1—figure supplement 1 contains IGV screenshots showing the pattern of split-reads characteristic of true- and false-positive non-reference TE insertions with TSDs.
DOI: http://dx.doi.org/10.7554/eLife.15716.002